
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000479649 | YWHAH-206 | 662 | - | - | - (aa) | - | - |
| ENST00000443669 | YWHAH-204 | 822 | - | ENSP00000408468 | 39 (aa) | - | F8WEB6 |
| ENST00000397492 | YWHAH-202 | 1879 | - | ENSP00000380629 | 72 (aa) | - | A2IDB1 |
| ENST00000248975 | YWHAH-201 | 1825 | - | ENSP00000248975 | 246 (aa) | - | Q04917 |
| ENST00000471374 | YWHAH-205 | 1735 | - | - | - (aa) | - | - |
| ENST00000420430 | YWHAH-203 | 502 | - | ENSP00000406747 | 162 (aa) | - | A2IDB2 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000128245 | rs149411586 | 22 | 31955305 | A | Post bronchodilator FEV1 | 26634245 | NR unit decrease | 0.25 | EFO_0004314|GO_0097366 |
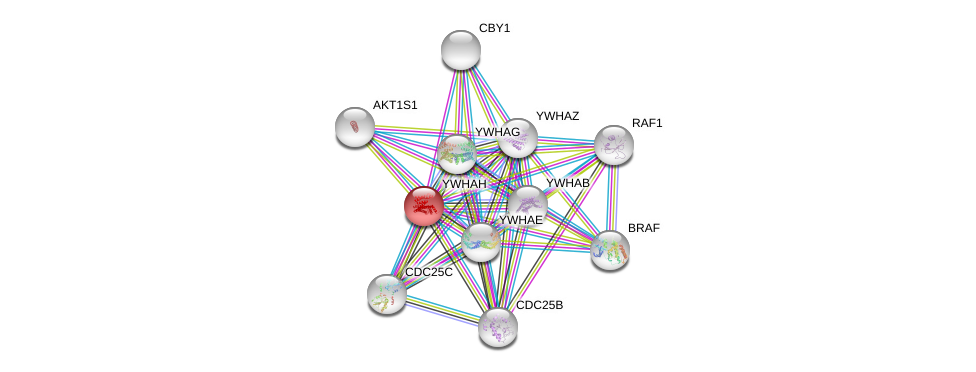
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000128245 | YWHAH | 100 | 86.992 | ENSG00000170027 | YWHAG | 99 | 86.992 | Homo_sapiens |
| ENSG00000128245 | YWHAH | 99 | 63.265 | ENSG00000108953 | YWHAE | 100 | 69.565 | Homo_sapiens |
| ENSG00000128245 | YWHAH | 100 | 99.187 | ENSMUSG00000018965 | Ywhah | 100 | 99.187 | Mus_musculus |
| ENSG00000128245 | YWHAH | 100 | 86.992 | ENSMUSG00000051391 | Ywhag | 99 | 86.992 | Mus_musculus |
| ENSG00000128245 | YWHAH | 99 | 63.265 | ENSMUSG00000020849 | Ywhae | 94 | 63.265 | Mus_musculus |
| ENSG00000128245 | YWHAH | 95 | 63.675 | YDR099W | BMH2 | 85 | 63.675 | Saccharomyces_cerevisiae |
| ENSG00000128245 | YWHAH | 98 | 62.140 | YER177W | BMH1 | 87 | 63.675 | Saccharomyces_cerevisiae |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0002028 | regulation of sodium ion transport | 16728661. | IDA | Process |
| GO:0003779 | actin binding | - | IEA | Function |
| GO:0005159 | insulin-like growth factor receptor binding | - | ISS | Function |
| GO:0005515 | protein binding | 10713667.11697890.12524439.14676191.16545136.16672277.16775625.16959763.17979178.18045992.18242179.18397886.18458160.19172738.19553522.19615732.19640509.19860830.20642453.23075850.23572552.23622247.23752268.24895406.25852190.26496610. | IPI | Function |
| GO:0005737 | cytoplasm | - | ISS | Component |
| GO:0005739 | mitochondrion | - | IEA | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0005886 | plasma membrane | 16728661. | IDA | Component |
| GO:0006713 | glucocorticoid catabolic process | 15790729. | IDA | Process |
| GO:0006886 | intracellular protein transport | - | ISS | Process |
| GO:0014704 | intercalated disc | 16728661. | IC | Component |
| GO:0017080 | sodium channel regulator activity | 16728661. | IDA | Function |
| GO:0019899 | enzyme binding | 17255105. | IPI | Function |
| GO:0019904 | protein domain specific binding | - | ISS | Function |
| GO:0021762 | substantia nigra development | 22926577. | HEP | Process |
| GO:0035259 | glucocorticoid receptor binding | 9079630. | IPI | Function |
| GO:0042802 | identical protein binding | 17085597.17979178. | IPI | Function |
| GO:0042921 | glucocorticoid receptor signaling pathway | 15790729. | IDA | Process |
| GO:0044325 | ion channel binding | 11953308.16728661. | IPI | Function |
| GO:0045664 | regulation of neuron differentiation | - | ISS | Process |
| GO:0045893 | positive regulation of transcription, DNA-templated | 15790729. | IDA | Process |
| GO:0046982 | protein heterodimerization activity | 11953308. | IPI | Function |
| GO:0048167 | regulation of synaptic plasticity | - | ISS | Process |
| GO:0050774 | negative regulation of dendrite morphogenesis | - | ISS | Process |
| GO:0061024 | membrane organization | - | TAS | Process |
| GO:0070062 | extracellular exosome | 19056867.19199708.20458337. | HDA | Component |
| GO:0086010 | membrane depolarization during action potential | 16728661. | IDA | Process |
| GO:0098793 | presynapse | - | IEA | Component |
| GO:0098978 | glutamatergic synapse | - | IEA | Component |
| GO:1900740 | positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway | - | TAS | Process |
| GO:2000649 | regulation of sodium ion transmembrane transporter activity | 16728661. | IDA | Process |