
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000558978 | DUT-206 | 1171 | - | ENSP00000452814 | 226 (aa) | - | H0YKI0 |
| ENST00000559416 | DUT-207 | 635 | - | ENSP00000454183 | 167 (aa) | - | H0YNW5 |
| ENST00000558813 | DUT-205 | 769 | - | ENSP00000453717 | 141 (aa) | - | A0A0C4DGL3 |
| ENST00000558367 | DUT-203 | 627 | - | ENSP00000453683 | 107 (aa) | - | H0YMP1 |
| ENST00000559852 | DUT-209 | 340 | - | - | - (aa) | - | - |
| ENST00000561350 | DUT-211 | 201 | - | - | - (aa) | - | - |
| ENST00000558472 | DUT-204 | 724 | - | ENSP00000452749 | 221 (aa) | - | H0YKC5 |
| ENST00000559540 | DUT-208 | 1441 | - | ENSP00000454041 | 143 (aa) | - | H0YNJ9 |
| ENST00000559935 | DUT-210 | 569 | - | ENSP00000453667 | 133 (aa) | - | H0YMM5 |
| ENST00000331200 | DUT-201 | 2147 | - | ENSP00000370376 | 252 (aa) | - | P33316 |
| ENST00000455976 | DUT-202 | 1935 | - | ENSP00000405160 | 164 (aa) | - | P33316 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000128951 | rs11637235 | 15 | 48340956 | ? | Protein quantitative trait loci | 18464913 | EFO_0004502 |
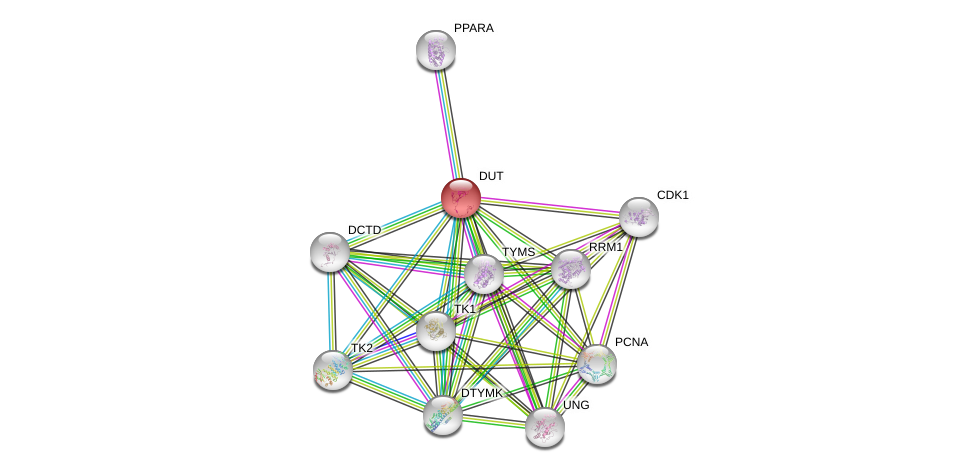
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000128951 | DUT | 99 | 92.143 | ENSMUSG00000027203 | Dut | 100 | 86.585 | Mus_musculus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000287 | magnesium ion binding | 21873635. | IBA | Function |
| GO:0003723 | RNA binding | 22658674. | HDA | Function |
| GO:0004170 | dUTP diphosphatase activity | 21873635. | IBA | Function |
| GO:0005515 | protein binding | 16189514.19060904. | IPI | Function |
| GO:0005634 | nucleus | 8631816. | TAS | Component |
| GO:0005654 | nucleoplasm | - | IDA | Component |
| GO:0005654 | nucleoplasm | - | TAS | Component |
| GO:0005739 | mitochondrion | - | IEA | Component |
| GO:0006139 | nucleobase-containing compound metabolic process | 1325640. | TAS | Process |
| GO:0006226 | dUMP biosynthetic process | 21873635. | IBA | Process |
| GO:0006226 | dUMP biosynthetic process | - | IEA | Process |
| GO:0006260 | DNA replication | 1325640. | TAS | Process |
| GO:0015949 | nucleobase-containing small molecule interconversion | - | TAS | Process |
| GO:0046081 | dUTP catabolic process | 21873635. | IBA | Process |
| GO:0070062 | extracellular exosome | 20458337. | HDA | Component |