
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000409382 | MMR_HSR1 | PF01926.23 | 1.8e-06 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000535507 | CRACR2A-206 | 2511 | - | - | - (aa) | - | - |
| ENST00000333750 | CRACR2A-202 | 3207 | - | ENSP00000331047 | 198 (aa) | - | H7BXT2 |
| ENST00000440314 | CRACR2A-203 | 2697 | XM_006719021 | ENSP00000409382 | 731 (aa) | XP_006719084 | Q9BSW2 |
| ENST00000514026 | CRACR2A-204 | 7554 | - | - | - (aa) | - | - |
| ENST00000535292 | CRACR2A-205 | 630 | - | ENSP00000438777 | 118 (aa) | - | H0YFI5 |
| ENST00000252322 | CRACR2A-201 | 2186 | - | ENSP00000252322 | 395 (aa) | - | Q9BSW2 |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000130038 | Parkinson Disease | 3.734896E-5 | 17052657 |
| ENSG00000130038 | Myocardial Infarction | 6.1480995E-004 | - |
| ENSG00000130038 | Body Height | 4.813e-006 | 18193045 |
| ENSG00000130038 | Body Height | 3.239e-006 | 18193045 |
| ENSG00000130038 | Altruism | 5.468e-005 | - |
| ENSG00000130038 | Ankle Brachial Index | 9.9556637E-005 | - |
| ENSG00000130038 | Platelet Function Tests | 9.8800000E-006 | - |
| ENSG00000130038 | Platelet Function Tests | 4.5700000E-006 | - |
| ENSG00000130038 | Parathyroid Hormone | 6.4860000E-005 | - |
| ENSG00000130038 | Breast Neoplasms | 5.5100000E-006 | - |
| ENSG00000130038 | Breast Neoplasms | 2.1990000E-005 | - |
| ENSG00000130038 | Breast Neoplasms | 5.1320000E-005 | - |
| ENSG00000130038 | Neuropsychological Tests | 8E-6 | 20125193 |
| ENSG00000130038 | Insulin Resistance | 9E-6 | 24204828 |
| ENSG00000130038 | Insulin | 8E-6 | 24204828 |
| ENSG00000130038 | Liver Cirrhosis | 8E-7 | 20708005 |
| ENSG00000130038 | Non-alcoholic Fatty Liver Disease | 8E-7 | 20708005 |
| ENSG00000130038 | Infertility, Male | 4E-6 | 19478329 |
| ENSG00000130038 | Viruses | 5E-6 | 21993531 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000130038 | rs10848918 | 12 | 3744163 | ? | Velopharyngeal dysfunction | 29855589 | [NR] unit decrease | 0.11454327 | EFO_0009336 |
| ENSG00000130038 | rs10744625 | 12 | 3759002 | C | Homeostasis model assessment of insulin resistance (dietary factor interaction) | 24204828 | EFO_0008111|EFO_0004501 | ||
| ENSG00000130038 | rs10744625 | 12 | 3759002 | C | Fasting insulin (dietary factor interaction) | 24204828 | EFO_0008111|EFO_0004466 | ||
| ENSG00000130038 | rs10848911 | 12 | 3705072 | ? | Male infertility | 19478329 | EFO_0004248 | ||
| ENSG00000130038 | rs10744620 | 12 | 3629928 | C | Monocyte chemoattractant protein-1 levels | 27989323 | [0.047-0.11] SD units decrease | 0.0788 | EFO_0004749 |
| ENSG00000130038 | rs4766152 | 12 | 3640962 | G | Cytomegalovirus antibody response | 21993531 | [0.13-0.32] unit decrease | 0.223 | GO_0009615 |
| ENSG00000130038 | rs887304 | 12 | 3648382 | A | Non-alcoholic fatty liver disease histology (lobular) | 20708005 | [NR] unit increase | 0.36 | EFO_0003095|EFO_0001422 |
| ENSG00000130038 | rs242016 | 12 | 3679094 | A | Chronic periodontitis (localised) | 30284742 | [2.32-6.29] | 3.7 | EFO_0000649 |
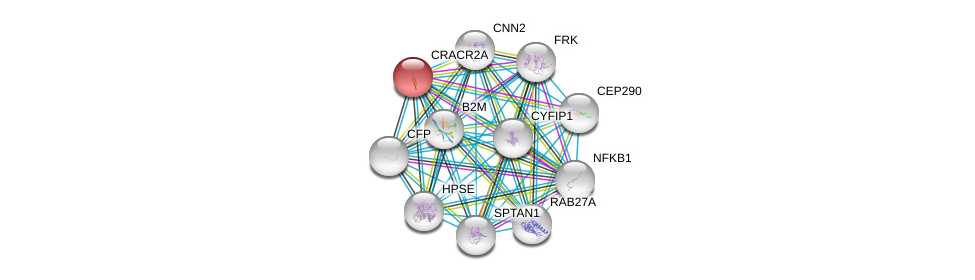
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000130038 | CRACR2A | 83 | 46.939 | ENSAPOG00000009642 | cracr2b | 93 | 45.238 | Acanthochromis_polyacanthus |
| ENSG00000130038 | CRACR2A | 91 | 61.667 | ENSAPOG00000013779 | cracr2ab | 83 | 57.778 | Acanthochromis_polyacanthus |
| ENSG00000130038 | CRACR2A | 78 | 88.312 | ENSAMEG00000005041 | - | 97 | 86.747 | Ailuropoda_melanoleuca |
| ENSG00000130038 | CRACR2A | 91 | 61.667 | ENSACIG00000010099 | CRACR2A | 94 | 54.131 | Amphilophus_citrinellus |
| ENSG00000130038 | CRACR2A | 88 | 62.857 | ENSACIG00000004009 | cracr2ab | 97 | 50.771 | Amphilophus_citrinellus |
| ENSG00000130038 | CRACR2A | 91 | 61.667 | ENSAOCG00000023189 | cracr2ab | 81 | 58.601 | Amphiprion_ocellaris |
| ENSG00000130038 | CRACR2A | 91 | 62.778 | ENSAOCG00000008147 | - | 94 | 55.274 | Amphiprion_ocellaris |
| ENSG00000130038 | CRACR2A | 84 | 47.619 | ENSAOCG00000019493 | cracr2b | 92 | 47.619 | Amphiprion_ocellaris |
| ENSG00000130038 | CRACR2A | 85 | 46.701 | ENSAPEG00000021843 | cracr2b | 94 | 46.701 | Amphiprion_percula |
| ENSG00000130038 | CRACR2A | 91 | 62.778 | ENSAPEG00000002244 | CRACR2A | 94 | 54.965 | Amphiprion_percula |
| ENSG00000130038 | CRACR2A | 88 | 62.857 | ENSAPEG00000006383 | cracr2ab | 99 | 48.753 | Amphiprion_percula |
| ENSG00000130038 | CRACR2A | 86 | 45.495 | ENSATEG00000010344 | cracr2b | 96 | 45.495 | Anabas_testudineus |
| ENSG00000130038 | CRACR2A | 91 | 61.111 | ENSATEG00000018003 | cracr2ab | 81 | 57.602 | Anabas_testudineus |
| ENSG00000130038 | CRACR2A | 91 | 62.222 | ENSATEG00000017411 | CRACR2A | 98 | 53.444 | Anabas_testudineus |
| ENSG00000130038 | CRACR2A | 93 | 41.126 | ENSACAG00000015413 | CRACR2B | 94 | 40.693 | Anolis_carolinensis |
| ENSG00000130038 | CRACR2A | 100 | 93.570 | ENSANAG00000036979 | CRACR2A | 100 | 93.570 | Aotus_nancymaae |
| ENSG00000130038 | CRACR2A | 91 | 61.111 | ENSACLG00000023670 | cracr2ab | 81 | 57.895 | Astatotilapia_calliptera |
| ENSG00000130038 | CRACR2A | 91 | 63.889 | ENSACLG00000007547 | CRACR2A | 94 | 54.986 | Astatotilapia_calliptera |
| ENSG00000130038 | CRACR2A | 91 | 34.574 | ENSACLG00000013916 | cracr2b | 89 | 43.939 | Astatotilapia_calliptera |
| ENSG00000130038 | CRACR2A | 58 | 46.137 | ENSAMXG00000010368 | cracr2ab | 99 | 46.358 | Astyanax_mexicanus |
| ENSG00000130038 | CRACR2A | 100 | 82.787 | ENSBTAG00000018940 | CRACR2A | 100 | 82.650 | Bos_taurus |
| ENSG00000130038 | CRACR2A | 100 | 92.202 | ENSCJAG00000018917 | CRACR2A | 100 | 92.202 | Callithrix_jacchus |
| ENSG00000130038 | CRACR2A | 100 | 82.377 | ENSCAFG00000015404 | CRACR2A | 100 | 82.104 | Canis_familiaris |
| ENSG00000130038 | CRACR2A | 100 | 82.377 | ENSCAFG00020014835 | CRACR2A | 100 | 82.104 | Canis_lupus_dingo |
| ENSG00000130038 | CRACR2A | 100 | 82.377 | ENSCHIG00000018141 | CRACR2A | 100 | 82.377 | Capra_hircus |
| ENSG00000130038 | CRACR2A | 100 | 86.749 | ENSTSYG00000014194 | CRACR2A | 100 | 86.749 | Carlito_syrichta |
| ENSG00000130038 | CRACR2A | 86 | 77.229 | ENSCAPG00000006188 | CRACR2A | 100 | 77.229 | Cavia_aperea |
| ENSG00000130038 | CRACR2A | 99 | 81.669 | ENSCPOG00000020119 | CRACR2A | 99 | 81.669 | Cavia_porcellus |
| ENSG00000130038 | CRACR2A | 100 | 92.066 | ENSCCAG00000019735 | CRACR2A | 100 | 92.066 | Cebus_capucinus |
| ENSG00000130038 | CRACR2A | 100 | 96.990 | ENSCATG00000040964 | CRACR2A | 100 | 96.990 | Cercocebus_atys |
| ENSG00000130038 | CRACR2A | 100 | 80.628 | ENSCLAG00000013246 | CRACR2A | 100 | 80.628 | Chinchilla_lanigera |
| ENSG00000130038 | CRACR2A | 100 | 96.322 | ENSCSAG00000011199 | CRACR2A | 100 | 96.322 | Chlorocebus_sabaeus |
| ENSG00000130038 | CRACR2A | 88 | 82.184 | ENSCPBG00000009731 | CRACR2A | 99 | 64.218 | Chrysemys_picta_bellii |
| ENSG00000130038 | CRACR2A | 93 | 42.877 | ENSCPBG00000007146 | CRACR2B | 73 | 56.019 | Chrysemys_picta_bellii |
| ENSG00000130038 | CRACR2A | 100 | 96.717 | ENSCANG00000032776 | CRACR2A | 100 | 96.717 | Colobus_angolensis_palliatus |
| ENSG00000130038 | CRACR2A | 100 | 80.191 | ENSCGRG00001008752 | - | 100 | 80.191 | Cricetulus_griseus_chok1gshd |
| ENSG00000130038 | CRACR2A | 100 | 79.508 | ENSCGRG00000015501 | - | 100 | 79.508 | Cricetulus_griseus_crigri |
| ENSG00000130038 | CRACR2A | 91 | 55.000 | ENSCSEG00000003612 | CRACR2A | 93 | 50.575 | Cynoglossus_semilaevis |
| ENSG00000130038 | CRACR2A | 96 | 56.701 | ENSCSEG00000008307 | cracr2ab | 80 | 55.263 | Cynoglossus_semilaevis |
| ENSG00000130038 | CRACR2A | 78 | 45.745 | ENSCSEG00000007618 | cracr2b | 94 | 45.128 | Cynoglossus_semilaevis |
| ENSG00000130038 | CRACR2A | 91 | 57.778 | ENSCVAG00000009954 | cracr2ab | 97 | 51.893 | Cyprinodon_variegatus |
| ENSG00000130038 | CRACR2A | 90 | 59.218 | ENSCVAG00000013782 | CRACR2A | 97 | 52.849 | Cyprinodon_variegatus |
| ENSG00000130038 | CRACR2A | 90 | 59.218 | ENSDARG00000068231 | CRACR2A | 96 | 54.232 | Danio_rerio |
| ENSG00000130038 | CRACR2A | 90 | 56.983 | ENSDARG00000045758 | cracr2ab | 99 | 46.737 | Danio_rerio |
| ENSG00000130038 | CRACR2A | 100 | 77.973 | ENSDNOG00000043722 | CRACR2A | 99 | 77.973 | Dasypus_novemcinctus |
| ENSG00000130038 | CRACR2A | 100 | 83.060 | ENSEASG00005017095 | CRACR2A | 100 | 83.060 | Equus_asinus_asinus |
| ENSG00000130038 | CRACR2A | 100 | 84.563 | ENSECAG00000034127 | CRACR2A | 100 | 84.563 | Equus_caballus |
| ENSG00000130038 | CRACR2A | 90 | 59.218 | ENSELUG00000018918 | CRACR2A | 94 | 53.249 | Esox_lucius |
| ENSG00000130038 | CRACR2A | 90 | 58.101 | ENSELUG00000001781 | cracr2ab | 73 | 56.176 | Esox_lucius |
| ENSG00000130038 | CRACR2A | 100 | 81.967 | ENSFCAG00000015741 | CRACR2A | 100 | 81.967 | Felis_catus |
| ENSG00000130038 | CRACR2A | 88 | 81.609 | ENSFALG00000004782 | CRACR2A | 99 | 63.534 | Ficedula_albicollis |
| ENSG00000130038 | CRACR2A | 100 | 77.467 | ENSFDAG00000013877 | CRACR2A | 100 | 77.467 | Fukomys_damarensis |
| ENSG00000130038 | CRACR2A | 91 | 59.444 | ENSGAFG00000008771 | cracr2ab | 97 | 47.125 | Gambusia_affinis |
| ENSG00000130038 | CRACR2A | 89 | 35.977 | ENSGAFG00000020778 | cracr2b | 81 | 46.842 | Gambusia_affinis |
| ENSG00000130038 | CRACR2A | 88 | 60.345 | ENSGAFG00000014607 | CRACR2A | 94 | 52.496 | Gambusia_affinis |
| ENSG00000130038 | CRACR2A | 88 | 81.609 | ENSGAGG00000002264 | CRACR2A | 99 | 64.315 | Gopherus_agassizii |
| ENSG00000130038 | CRACR2A | 100 | 99.590 | ENSGGOG00000010289 | CRACR2A | 100 | 99.590 | Gorilla_gorilla |
| ENSG00000130038 | CRACR2A | 91 | 61.111 | ENSHBUG00000018719 | cracr2ab | 81 | 57.895 | Haplochromis_burtoni |
| ENSG00000130038 | CRACR2A | 91 | 63.333 | ENSHBUG00000017137 | CRACR2A | 94 | 54.843 | Haplochromis_burtoni |
| ENSG00000130038 | CRACR2A | 100 | 79.673 | ENSHGLG00000003776 | CRACR2A | 100 | 79.673 | Heterocephalus_glaber_female |
| ENSG00000130038 | CRACR2A | 100 | 79.564 | ENSHGLG00100005482 | CRACR2A | 100 | 79.564 | Heterocephalus_glaber_male |
| ENSG00000130038 | CRACR2A | 94 | 56.061 | ENSHCOG00000004942 | cracr2ab | 97 | 50.644 | Hippocampus_comes |
| ENSG00000130038 | CRACR2A | 91 | 58.889 | ENSHCOG00000006842 | CRACR2A | 99 | 51.626 | Hippocampus_comes |
| ENSG00000130038 | CRACR2A | 91 | 56.111 | ENSIPUG00000017914 | cracr2ab | 94 | 48.237 | Ictalurus_punctatus |
| ENSG00000130038 | CRACR2A | 99 | 53.042 | ENSIPUG00000015191 | CRACR2A | 100 | 52.722 | Ictalurus_punctatus |
| ENSG00000130038 | CRACR2A | 81 | 46.429 | ENSIPUG00000013598 | cracr2b | 90 | 46.429 | Ictalurus_punctatus |
| ENSG00000130038 | CRACR2A | 100 | 85.130 | ENSSTOG00000021197 | CRACR2A | 99 | 85.130 | Ictidomys_tridecemlineatus |
| ENSG00000130038 | CRACR2A | 97 | 74.022 | ENSJJAG00000014186 | - | 98 | 74.022 | Jaculus_jaculus |
| ENSG00000130038 | CRACR2A | 90 | 60.894 | ENSKMAG00000021906 | CRACR2A | 96 | 51.136 | Kryptolebias_marmoratus |
| ENSG00000130038 | CRACR2A | 91 | 58.889 | ENSKMAG00000016358 | cracr2ab | 82 | 56.357 | Kryptolebias_marmoratus |
| ENSG00000130038 | CRACR2A | 99 | 32.527 | ENSLBEG00000014261 | cracr2b | 99 | 40.562 | Labrus_bergylta |
| ENSG00000130038 | CRACR2A | 88 | 60.920 | ENSLBEG00000018669 | cracr2ab | 97 | 50.431 | Labrus_bergylta |
| ENSG00000130038 | CRACR2A | 91 | 61.111 | ENSLBEG00000010713 | CRACR2A | 96 | 54.126 | Labrus_bergylta |
| ENSG00000130038 | CRACR2A | 93 | 41.270 | ENSLACG00000007778 | - | 100 | 39.633 | Latimeria_chalumnae |
| ENSG00000130038 | CRACR2A | 93 | 39.744 | ENSLACG00000009026 | - | 100 | 39.745 | Latimeria_chalumnae |
| ENSG00000130038 | CRACR2A | 91 | 62.222 | ENSLOCG00000016848 | cracr2ab | 96 | 56.838 | Lepisosteus_oculatus |
| ENSG00000130038 | CRACR2A | 100 | 96.443 | ENSMFAG00000036067 | CRACR2A | 100 | 96.443 | Macaca_fascicularis |
| ENSG00000130038 | CRACR2A | 100 | 96.306 | ENSMMUG00000023583 | CRACR2A | 100 | 96.306 | Macaca_mulatta |
| ENSG00000130038 | CRACR2A | 100 | 96.443 | ENSMNEG00000031773 | CRACR2A | 100 | 96.443 | Macaca_nemestrina |
| ENSG00000130038 | CRACR2A | 100 | 96.854 | ENSMLEG00000044159 | CRACR2A | 99 | 96.854 | Mandrillus_leucophaeus |
| ENSG00000130038 | CRACR2A | 91 | 59.444 | ENSMAMG00000011388 | cracr2ab | 82 | 56.598 | Mastacembelus_armatus |
| ENSG00000130038 | CRACR2A | 88 | 35.429 | ENSMAMG00000005360 | cracr2b | 88 | 43.694 | Mastacembelus_armatus |
| ENSG00000130038 | CRACR2A | 91 | 61.667 | ENSMAMG00000001270 | CRACR2A | 95 | 55.274 | Mastacembelus_armatus |
| ENSG00000130038 | CRACR2A | 88 | 62.286 | ENSMZEG00005005975 | cracr2ab | 96 | 50.509 | Maylandia_zebra |
| ENSG00000130038 | CRACR2A | 91 | 63.333 | ENSMZEG00005016858 | CRACR2A | 94 | 54.843 | Maylandia_zebra |
| ENSG00000130038 | CRACR2A | 100 | 79.945 | ENSMAUG00000010171 | - | 100 | 79.945 | Mesocricetus_auratus |
| ENSG00000130038 | CRACR2A | 100 | 86.066 | ENSMICG00000012907 | CRACR2A | 100 | 86.066 | Microcebus_murinus |
| ENSG00000130038 | CRACR2A | 99 | 80.301 | ENSMOCG00000016052 | - | 99 | 80.301 | Microtus_ochrogaster |
| ENSG00000130038 | CRACR2A | 91 | 60.000 | ENSMMOG00000019358 | CRACR2A | 94 | 52.045 | Mola_mola |
| ENSG00000130038 | CRACR2A | 91 | 60.556 | ENSMMOG00000001929 | cracr2ab | 97 | 50.491 | Mola_mola |
| ENSG00000130038 | CRACR2A | 91 | 59.444 | ENSMALG00000021818 | CRACR2A | 95 | 54.178 | Monopterus_albus |
| ENSG00000130038 | CRACR2A | 91 | 59.444 | ENSMALG00000012964 | cracr2ab | 78 | 55.556 | Monopterus_albus |
| ENSG00000130038 | CRACR2A | 93 | 35.584 | ENSMALG00000006310 | cracr2b | 99 | 42.623 | Monopterus_albus |
| ENSG00000130038 | CRACR2A | 100 | 80.328 | ENSMUSG00000061414 | Cracr2a | 100 | 80.328 | Mus_musculus |
| ENSG00000130038 | CRACR2A | 100 | 81.037 | ENSMPUG00000009336 | CRACR2A | 100 | 80.764 | Mustela_putorius_furo |
| ENSG00000130038 | CRACR2A | 100 | 81.855 | ENSNGAG00000014625 | - | 100 | 81.855 | Nannospalax_galili |
| ENSG00000130038 | CRACR2A | 88 | 62.286 | ENSNBRG00000005924 | cracr2ab | 96 | 50.796 | Neolamprologus_brichardi |
| ENSG00000130038 | CRACR2A | 91 | 62.222 | ENSNBRG00000015888 | CRACR2A | 91 | 59.327 | Neolamprologus_brichardi |
| ENSG00000130038 | CRACR2A | 100 | 95.622 | ENSNLEG00000003001 | CRACR2A | 99 | 95.622 | Nomascus_leucogenys |
| ENSG00000130038 | CRACR2A | 100 | 81.421 | ENSODEG00000002836 | CRACR2A | 100 | 81.421 | Octodon_degus |
| ENSG00000130038 | CRACR2A | 100 | 83.607 | ENSOCUG00000005317 | CRACR2A | 100 | 82.650 | Oryctolagus_cuniculus |
| ENSG00000130038 | CRACR2A | 91 | 58.889 | ENSORLG00000014854 | cracr2ab | 83 | 54.839 | Oryzias_latipes |
| ENSG00000130038 | CRACR2A | 91 | 59.444 | ENSORLG00000025905 | CRACR2A | 94 | 54.155 | Oryzias_latipes |
| ENSG00000130038 | CRACR2A | 91 | 60.000 | ENSORLG00020010617 | CRACR2A | 94 | 54.077 | Oryzias_latipes_hni |
| ENSG00000130038 | CRACR2A | 90 | 58.659 | ENSORLG00015013042 | cracr2ab | 83 | 54.545 | Oryzias_latipes_hsok |
| ENSG00000130038 | CRACR2A | 91 | 58.889 | ENSORLG00015001413 | CRACR2A | 98 | 50.860 | Oryzias_latipes_hsok |
| ENSG00000130038 | CRACR2A | 92 | 57.513 | ENSOMEG00000007213 | cracr2ab | 97 | 50.363 | Oryzias_melastigma |
| ENSG00000130038 | CRACR2A | 96 | 88.462 | ENSOGAG00000001502 | CRACR2A | 100 | 87.607 | Otolemur_garnettii |
| ENSG00000130038 | CRACR2A | 92 | 35.673 | ENSOARG00000013673 | - | 96 | 35.912 | Ovis_aries |
| ENSG00000130038 | CRACR2A | 100 | 81.992 | ENSOARG00000010465 | CRACR2A | 100 | 81.992 | Ovis_aries |
| ENSG00000130038 | CRACR2A | 100 | 99.180 | ENSPPAG00000026014 | CRACR2A | 100 | 99.180 | Pan_paniscus |
| ENSG00000130038 | CRACR2A | 100 | 81.557 | ENSPPRG00000006970 | CRACR2A | 100 | 81.557 | Panthera_pardus |
| ENSG00000130038 | CRACR2A | 100 | 99.180 | ENSPTRG00000004545 | CRACR2A | 100 | 99.180 | Pan_troglodytes |
| ENSG00000130038 | CRACR2A | 99 | 96.810 | ENSPANG00000025412 | CRACR2A | 98 | 96.810 | Papio_anubis |
| ENSG00000130038 | CRACR2A | 90 | 59.218 | ENSPKIG00000019679 | - | 95 | 51.748 | Paramormyrops_kingsleyae |
| ENSG00000130038 | CRACR2A | 98 | 50.344 | ENSPKIG00000001449 | - | 99 | 50.137 | Paramormyrops_kingsleyae |
| ENSG00000130038 | CRACR2A | 88 | 61.714 | ENSPMGG00000005501 | - | 90 | 52.496 | Periophthalmus_magnuspinnatus |
| ENSG00000130038 | CRACR2A | 91 | 60.556 | ENSPMGG00000009818 | cracr2ab | 97 | 52.540 | Periophthalmus_magnuspinnatus |
| ENSG00000130038 | CRACR2A | 66 | 76.923 | ENSPCIG00000001087 | - | 99 | 71.429 | Phascolarctos_cinereus |
| ENSG00000130038 | CRACR2A | 100 | 34.127 | ENSPFOG00000010947 | cracr2b | 90 | 45.789 | Poecilia_formosa |
| ENSG00000130038 | CRACR2A | 91 | 60.556 | ENSPFOG00000005266 | cracr2ab | 99 | 52.533 | Poecilia_formosa |
| ENSG00000130038 | CRACR2A | 94 | 35.233 | ENSPLAG00000003773 | cracr2b | 90 | 45.789 | Poecilia_latipinna |
| ENSG00000130038 | CRACR2A | 90 | 59.777 | ENSPLAG00000001077 | - | 94 | 54.196 | Poecilia_latipinna |
| ENSG00000130038 | CRACR2A | 91 | 60.000 | ENSPLAG00000003905 | cracr2ab | 97 | 51.778 | Poecilia_latipinna |
| ENSG00000130038 | CRACR2A | 83 | 46.316 | ENSPMEG00000020754 | cracr2b | 90 | 46.316 | Poecilia_mexicana |
| ENSG00000130038 | CRACR2A | 91 | 60.556 | ENSPMEG00000007308 | cracr2ab | 97 | 51.804 | Poecilia_mexicana |
| ENSG00000130038 | CRACR2A | 90 | 59.777 | ENSPMEG00000005485 | - | 94 | 54.054 | Poecilia_mexicana |
| ENSG00000130038 | CRACR2A | 90 | 59.218 | ENSPREG00000021082 | CRACR2A | 96 | 52.528 | Poecilia_reticulata |
| ENSG00000130038 | CRACR2A | 91 | 60.556 | ENSPREG00000002593 | cracr2ab | 97 | 52.542 | Poecilia_reticulata |
| ENSG00000130038 | CRACR2A | 100 | 87.158 | ENSPCOG00000020329 | CRACR2A | 100 | 87.158 | Propithecus_coquereli |
| ENSG00000130038 | CRACR2A | 91 | 63.333 | ENSPNYG00000020714 | CRACR2A | 92 | 57.851 | Pundamilia_nyererei |
| ENSG00000130038 | CRACR2A | 88 | 61.714 | ENSPNYG00000000606 | cracr2ab | 97 | 50.211 | Pundamilia_nyererei |
| ENSG00000130038 | CRACR2A | 90 | 60.894 | ENSPNAG00000020348 | CRACR2A | 94 | 55.651 | Pygocentrus_nattereri |
| ENSG00000130038 | CRACR2A | 91 | 57.778 | ENSPNAG00000029103 | cracr2ab | 96 | 49.591 | Pygocentrus_nattereri |
| ENSG00000130038 | CRACR2A | 100 | 96.990 | ENSRBIG00000044914 | CRACR2A | 100 | 96.990 | Rhinopithecus_bieti |
| ENSG00000130038 | CRACR2A | 88 | 98.857 | ENSRROG00000028125 | CRACR2A | 99 | 91.447 | Rhinopithecus_roxellana |
| ENSG00000130038 | CRACR2A | 100 | 92.760 | ENSSBOG00000024718 | CRACR2A | 100 | 92.760 | Saimiri_boliviensis_boliviensis |
| ENSG00000130038 | CRACR2A | 91 | 60.556 | ENSSFOG00015008628 | - | 98 | 53.024 | Scleropages_formosus |
| ENSG00000130038 | CRACR2A | 91 | 57.778 | ENSSFOG00015004037 | cracr2ab | 97 | 53.890 | Scleropages_formosus |
| ENSG00000130038 | CRACR2A | 93 | 35.580 | ENSSMAG00000013579 | cracr2b | 91 | 47.475 | Scophthalmus_maximus |
| ENSG00000130038 | CRACR2A | 91 | 58.889 | ENSSMAG00000001304 | cracr2ab | 81 | 55.942 | Scophthalmus_maximus |
| ENSG00000130038 | CRACR2A | 91 | 62.222 | ENSSMAG00000011200 | CRACR2A | 95 | 54.930 | Scophthalmus_maximus |
| ENSG00000130038 | CRACR2A | 91 | 62.222 | ENSSDUG00000000911 | CRACR2A | 95 | 53.547 | Seriola_dumerili |
| ENSG00000130038 | CRACR2A | 93 | 35.967 | ENSSDUG00000013283 | cracr2b | 93 | 43.243 | Seriola_dumerili |
| ENSG00000130038 | CRACR2A | 91 | 62.222 | ENSSLDG00000010742 | CRACR2A | 95 | 53.547 | Seriola_lalandi_dorsalis |
| ENSG00000130038 | CRACR2A | 88 | 61.714 | ENSSLDG00000023518 | cracr2ab | 94 | 48.798 | Seriola_lalandi_dorsalis |
| ENSG00000130038 | CRACR2A | 93 | 36.240 | ENSSLDG00000022995 | cracr2b | 89 | 46.632 | Seriola_lalandi_dorsalis |
| ENSG00000130038 | CRACR2A | 72 | 55.789 | ENSSPUG00000010462 | - | 59 | 50.917 | Sphenodon_punctatus |
| ENSG00000130038 | CRACR2A | 89 | 79.096 | ENSSPUG00000017268 | CRACR2A | 95 | 61.966 | Sphenodon_punctatus |
| ENSG00000130038 | CRACR2A | 91 | 62.222 | ENSSPAG00000012064 | CRACR2A | 94 | 55.367 | Stegastes_partitus |
| ENSG00000130038 | CRACR2A | 91 | 61.667 | ENSSPAG00000022330 | cracr2ab | 99 | 47.928 | Stegastes_partitus |
| ENSG00000130038 | CRACR2A | 98 | 83.357 | ENSSSCG00000000732 | CRACR2A | 100 | 84.897 | Sus_scrofa |
| ENSG00000130038 | CRACR2A | 99 | 55.238 | ENSTRUG00000023284 | CRACR2A | 92 | 53.957 | Takifugu_rubripes |
| ENSG00000130038 | CRACR2A | 100 | 80.874 | ENSUAMG00000021584 | CRACR2A | 100 | 80.601 | Ursus_americanus |
| ENSG00000130038 | CRACR2A | 100 | 80.601 | ENSUMAG00000014780 | CRACR2A | 100 | 80.328 | Ursus_maritimus |
| ENSG00000130038 | CRACR2A | 100 | 82.923 | ENSVVUG00000018097 | CRACR2A | 100 | 82.650 | Vulpes_vulpes |
| ENSG00000130038 | CRACR2A | 88 | 59.770 | ENSXCOG00000007494 | CRACR2A | 96 | 51.631 | Xiphophorus_couchianus |
| ENSG00000130038 | CRACR2A | 91 | 60.000 | ENSXCOG00000013604 | cracr2ab | 97 | 52.096 | Xiphophorus_couchianus |
| ENSG00000130038 | CRACR2A | 90 | 58.659 | ENSXMAG00000021050 | CRACR2A | 94 | 53.333 | Xiphophorus_maculatus |
| ENSG00000130038 | CRACR2A | 91 | 59.444 | ENSXMAG00000011202 | cracr2ab | 97 | 51.020 | Xiphophorus_maculatus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0002115 | store-operated calcium entry | 20418871. | IMP | Process |
| GO:0002250 | adaptive immune response | - | IEA | Process |
| GO:0005509 | calcium ion binding | 20418871. | IDA | Function |
| GO:0005515 | protein binding | 20418871.23088713.25416956. | IPI | Function |
| GO:0005576 | extracellular region | - | TAS | Component |
| GO:0005737 | cytoplasm | 20418871. | IDA | Component |
| GO:0016020 | membrane | 19946888. | HDA | Component |
| GO:0032237 | activation of store-operated calcium channel activity | 20418871. | IMP | Process |
| GO:0035580 | specific granule lumen | - | TAS | Component |
| GO:0043312 | neutrophil degranulation | - | TAS | Process |
| GO:0051928 | positive regulation of calcium ion transport | 20418871. | IMP | Process |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| ACC | |||||||
| ACC | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRP | |||||||
| KIRP | |||||||
| LAML | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SARC | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| THCA | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| COAD | |||
| DLBC | |||
| PAAD | |||
| READ | |||
| TGCT | |||
| THYM |
| Cancer | P-value | Q-value |
|---|---|---|
| THYM | ||
| KIRC | ||
| UCS | ||
| SKCM | ||
| PRAD | ||
| GBM | ||
| LGG | ||
| LUAD | ||
| UVM |