
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000469151 | PWI | PF01480.17 | 4.7e-31 | 1 | 1 |
| ENSP00000471084 | PWI | PF01480.17 | 1.9e-29 | 1 | 1 |
| ENSP00000391430 | PWI | PF01480.17 | 2.2e-29 | 1 | 1 |
| ENSP00000326261 | PWI | PF01480.17 | 2.3e-29 | 1 | 1 |
| ENSP00000363510 | PWI | PF01480.17 | 2.4e-29 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000496882 | SRRM1-220 | 834 | - | - | - (aa) | - | - |
| ENST00000596378 | SRRM1-222 | 2314 | - | ENSP00000471084 | 681 (aa) | - | M0R088 |
| ENST00000479034 | SRRM1-212 | 2671 | - | - | - (aa) | - | - |
| ENST00000461768 | SRRM1-204 | 496 | - | - | - (aa) | - | - |
| ENST00000490543 | SRRM1-217 | 673 | - | - | - (aa) | - | - |
| ENST00000474843 | SRRM1-210 | 817 | - | - | - (aa) | - | - |
| ENST00000470243 | SRRM1-208 | 721 | - | - | - (aa) | - | - |
| ENST00000600523 | SRRM1-223 | 575 | - | ENSP00000471819 | 79 (aa) | - | M0R1E7 |
| ENST00000495561 | SRRM1-219 | 563 | - | - | - (aa) | - | - |
| ENST00000374389 | SRRM1-202 | 3110 | XM_005245717 | ENSP00000363510 | 913 (aa) | XP_005245774 | A9Z1X7 |
| ENST00000323848 | SRRM1-201 | 4011 | XM_005245721 | ENSP00000326261 | 904 (aa) | XP_005245778 | Q8IYB3 |
| ENST00000489130 | SRRM1-216 | 513 | - | - | - (aa) | - | - |
| ENST00000478890 | SRRM1-211 | 480 | - | - | - (aa) | - | - |
| ENST00000494934 | SRRM1-218 | 802 | - | - | - (aa) | - | - |
| ENST00000485441 | SRRM1-213 | 561 | - | - | - (aa) | - | - |
| ENST00000447431 | SRRM1-203 | 3691 | - | ENSP00000391430 | 820 (aa) | - | E9PCT1 |
| ENST00000462710 | SRRM1-205 | 2624 | - | - | - (aa) | - | - |
| ENST00000466910 | SRRM1-206 | 799 | - | - | - (aa) | - | - |
| ENST00000593639 | SRRM1-221 | 563 | - | ENSP00000469151 | 101 (aa) | - | M0QXG5 |
| ENST00000485541 | SRRM1-214 | 679 | - | - | - (aa) | - | - |
| ENST00000471693 | SRRM1-209 | 377 | - | - | - (aa) | - | - |
| ENST00000468822 | SRRM1-207 | 768 | - | - | - (aa) | - | - |
| ENST00000488317 | SRRM1-215 | 333 | - | - | - (aa) | - | - |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000133226 | rs143916866 | 1 | 24641046 | T | Male-pattern baldness | 28196072 | NR unit decrease | 0.15486 | Orphanet_79364 |
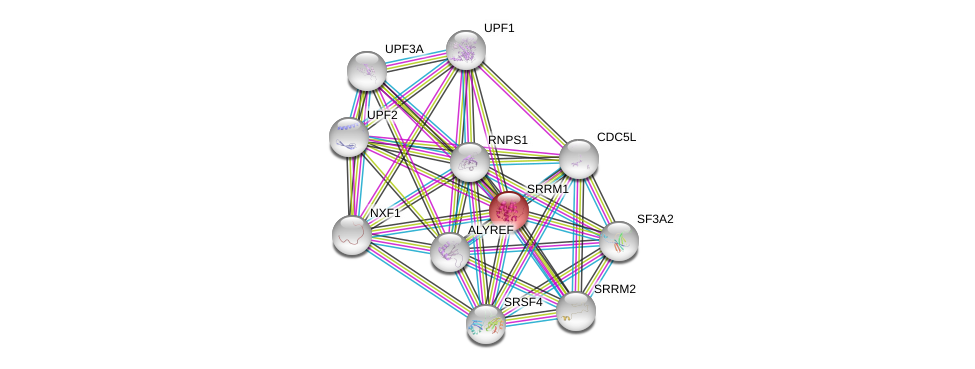
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000133226 | SRRM1 | 100 | 100.000 | ENSAMEG00000010075 | SRRM1 | 100 | 98.357 | Ailuropoda_melanoleuca |
| ENSG00000133226 | SRRM1 | 99 | 94.872 | ENSACIG00000023398 | - | 68 | 88.344 | Amphilophus_citrinellus |
| ENSG00000133226 | SRRM1 | 100 | 99.010 | ENSAPLG00000002066 | SRRM1 | 59 | 76.335 | Anas_platyrhynchos |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | ENSANAG00000035606 | SRRM1 | 100 | 99.004 | Aotus_nancymaae |
| ENSG00000133226 | SRRM1 | 99 | 94.872 | ENSACLG00000019786 | - | 68 | 88.272 | Astatotilapia_calliptera |
| ENSG00000133226 | SRRM1 | 100 | 94.059 | ENSAMXG00000040320 | - | 99 | 71.084 | Astyanax_mexicanus |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | ENSBTAG00000013772 | SRRM1 | 100 | 97.677 | Bos_taurus |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | ENSCJAG00000020822 | SRRM1 | 100 | 98.576 | Callithrix_jacchus |
| ENSG00000133226 | SRRM1 | 100 | 98.020 | ENSCAFG00020000003 | - | 98 | 89.573 | Canis_lupus_dingo |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | ENSCAFG00020020948 | SRRM1 | 100 | 98.230 | Canis_lupus_dingo |
| ENSG00000133226 | SRRM1 | 96 | 98.684 | ENSCHIG00000021159 | - | 54 | 98.684 | Capra_hircus |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | ENSCHIG00000014656 | SRRM1 | 100 | 96.507 | Capra_hircus |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | ENSTSYG00000006782 | SRRM1 | 100 | 98.357 | Carlito_syrichta |
| ENSG00000133226 | SRRM1 | 100 | 94.436 | ENSCAPG00000011473 | SRRM1 | 100 | 94.649 | Cavia_aperea |
| ENSG00000133226 | SRRM1 | 99 | 100.000 | ENSCPOG00000015594 | SRRM1 | 100 | 95.575 | Cavia_porcellus |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | ENSCCAG00000029474 | SRRM1 | 100 | 98.795 | Cebus_capucinus |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | ENSCATG00000031303 | SRRM1 | 100 | 99.558 | Cercocebus_atys |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | ENSCLAG00000014592 | SRRM1 | 100 | 96.571 | Chinchilla_lanigera |
| ENSG00000133226 | SRRM1 | 79 | 97.486 | ENSCHOG00000000027 | SRRM1 | 73 | 98.873 | Choloepus_hoffmanni |
| ENSG00000133226 | SRRM1 | 96 | 72.368 | ENSCING00000022242 | - | 94 | 66.443 | Ciona_intestinalis |
| ENSG00000133226 | SRRM1 | 96 | 77.632 | ENSCSAVG00000003919 | - | 77 | 68.098 | Ciona_savignyi |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | ENSCANG00000028271 | SRRM1 | 100 | 99.562 | Colobus_angolensis_palliatus |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | ENSCGRG00001014174 | Srrm1 | 77 | 95.779 | Cricetulus_griseus_chok1gshd |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | ENSCGRG00000017603 | Srrm1 | 75 | 95.779 | Cricetulus_griseus_crigri |
| ENSG00000133226 | SRRM1 | 99 | 93.590 | ENSCVAG00000000504 | - | 67 | 93.590 | Cyprinodon_variegatus |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | ENSDNOG00000049141 | SRRM1 | 98 | 93.100 | Dasypus_novemcinctus |
| ENSG00000133226 | SRRM1 | 99 | 100.000 | ENSDORG00000028601 | - | 100 | 82.456 | Dipodomys_ordii |
| ENSG00000133226 | SRRM1 | 99 | 100.000 | ENSETEG00000006817 | SRRM1 | 59 | 95.733 | Echinops_telfairi |
| ENSG00000133226 | SRRM1 | 100 | 80.198 | ENSEBUG00000002653 | - | 81 | 72.727 | Eptatretus_burgeri |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | ENSEASG00005004270 | SRRM1 | 100 | 96.623 | Equus_asinus_asinus |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | ENSECAG00000026981 | SRRM1 | 100 | 97.345 | Equus_caballus |
| ENSG00000133226 | SRRM1 | 99 | 100.000 | ENSEEUG00000013830 | SRRM1 | 63 | 90.385 | Erinaceus_europaeus |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | ENSFCAG00000000678 | - | 100 | 98.451 | Felis_catus |
| ENSG00000133226 | SRRM1 | 99 | 83.333 | ENSFCAG00000044640 | - | 73 | 83.750 | Felis_catus |
| ENSG00000133226 | SRRM1 | 100 | 99.010 | ENSFCAG00000034671 | - | 85 | 89.069 | Felis_catus |
| ENSG00000133226 | SRRM1 | 100 | 99.010 | ENSFALG00000000476 | SRRM1 | 100 | 100.000 | Ficedula_albicollis |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | ENSFDAG00000009903 | SRRM1 | 100 | 97.345 | Fukomys_damarensis |
| ENSG00000133226 | SRRM1 | 99 | 93.590 | ENSFHEG00000007359 | - | 83 | 91.358 | Fundulus_heteroclitus |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | ENSGALG00000001283 | SRRM1 | 100 | 100.000 | Gallus_gallus |
| ENSG00000133226 | SRRM1 | 99 | 94.872 | ENSGAFG00000019431 | - | 98 | 62.406 | Gambusia_affinis |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | ENSGGOG00000005209 | SRRM1 | 100 | 99.890 | Gorilla_gorilla |
| ENSG00000133226 | SRRM1 | 100 | 94.937 | ENSHBUG00000020354 | - | 100 | 94.937 | Haplochromis_burtoni |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | ENSHGLG00000017105 | SRRM1 | 100 | 97.345 | Heterocephalus_glaber_female |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | ENSHGLG00100016772 | SRRM1 | 99 | 96.889 | Heterocephalus_glaber_male |
| ENSG00000133226 | SRRM1 | 99 | 79.487 | ENSHGLG00100010430 | - | 99 | 78.750 | Heterocephalus_glaber_male |
| ENSG00000133226 | SRRM1 | 94 | 97.895 | ENSHGLG00100017918 | - | 100 | 90.299 | Heterocephalus_glaber_male |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | ENSSTOG00000014640 | SRRM1 | 100 | 98.562 | Ictidomys_tridecemlineatus |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | ENSJJAG00000018643 | Srrm1 | 74 | 93.558 | Jaculus_jaculus |
| ENSG00000133226 | SRRM1 | 100 | 93.671 | ENSKMAG00000013810 | - | 88 | 93.671 | Kryptolebias_marmoratus |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | ENSLAFG00000013830 | SRRM1 | 100 | 94.486 | Loxodonta_africana |
| ENSG00000133226 | SRRM1 | 99 | 72.000 | ENSMFAG00000038841 | - | 61 | 80.885 | Macaca_fascicularis |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | ENSMFAG00000025726 | SRRM1 | 100 | 99.668 | Macaca_fascicularis |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | ENSMMUG00000006645 | SRRM1 | 100 | 99.558 | Macaca_mulatta |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | ENSMNEG00000039415 | SRRM1 | 100 | 99.668 | Macaca_nemestrina |
| ENSG00000133226 | SRRM1 | 61 | 91.935 | ENSMLEG00000032221 | - | 61 | 73.764 | Mandrillus_leucophaeus |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | ENSMLEG00000038030 | - | 100 | 93.428 | Mandrillus_leucophaeus |
| ENSG00000133226 | SRRM1 | 99 | 94.872 | ENSMZEG00005006344 | - | 68 | 88.272 | Maylandia_zebra |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | ENSMICG00000006553 | SRRM1 | 100 | 98.451 | Microcebus_murinus |
| ENSG00000133226 | SRRM1 | 99 | 94.872 | ENSMMOG00000002390 | - | 77 | 94.872 | Mola_mola |
| ENSG00000133226 | SRRM1 | 100 | 92.079 | ENSMALG00000010813 | - | 67 | 76.687 | Monopterus_albus |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | MGP_CAROLIEiJ_G0026636 | Srrm1 | 100 | 85.965 | Mus_caroli |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | ENSMUSG00000028809 | Srrm1 | 100 | 85.965 | Mus_musculus |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | MGP_PahariEiJ_G0028969 | Srrm1 | 100 | 85.965 | Mus_pahari |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | MGP_SPRETEiJ_G0027616 | Srrm1 | 100 | 85.965 | Mus_spretus |
| ENSG00000133226 | SRRM1 | 100 | 98.020 | ENSMPUG00000000018 | - | 83 | 98.780 | Mustela_putorius_furo |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | ENSMPUG00000015831 | - | 99 | 87.552 | Mustela_putorius_furo |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | ENSNGAG00000014562 | Srrm1 | 75 | 95.362 | Nannospalax_galili |
| ENSG00000133226 | SRRM1 | 99 | 94.872 | ENSNBRG00000010911 | - | 67 | 87.730 | Neolamprologus_brichardi |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | ENSNLEG00000008966 | SRRM1 | 100 | 100.000 | Nomascus_leucogenys |
| ENSG00000133226 | SRRM1 | 71 | 80.000 | ENSMEUG00000014704 | SRRM1 | 57 | 83.598 | Notamacropus_eugenii |
| ENSG00000133226 | SRRM1 | 99 | 89.744 | ENSODEG00000002361 | - | 100 | 89.744 | Octodon_degus |
| ENSG00000133226 | SRRM1 | 96 | 93.421 | ENSODEG00000007221 | - | 99 | 93.421 | Octodon_degus |
| ENSG00000133226 | SRRM1 | 99 | 96.154 | ENSODEG00000019473 | - | 100 | 96.154 | Octodon_degus |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | ENSOCUG00000008438 | SRRM1 | 100 | 97.162 | Oryctolagus_cuniculus |
| ENSG00000133226 | SRRM1 | 99 | 94.872 | ENSORLG00000017510 | - | 68 | 87.117 | Oryzias_latipes |
| ENSG00000133226 | SRRM1 | 100 | 94.937 | ENSORLG00020018419 | - | 100 | 94.937 | Oryzias_latipes_hni |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | ENSOGAG00000010210 | SRRM1 | 100 | 96.950 | Otolemur_garnettii |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | ENSOARG00000006453 | SRRM1 | 99 | 97.406 | Ovis_aries |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | ENSPPAG00000043918 | SRRM1 | 100 | 99.890 | Pan_paniscus |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | ENSPPRG00000002509 | SRRM1 | 100 | 98.451 | Panthera_pardus |
| ENSG00000133226 | SRRM1 | 100 | 87.129 | ENSPTIG00000022093 | - | 74 | 87.261 | Panthera_tigris_altaica |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | ENSPTIG00000008778 | - | 100 | 92.896 | Panthera_tigris_altaica |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | ENSPTRG00000000349 | SRRM1 | 100 | 100.000 | Pan_troglodytes |
| ENSG00000133226 | SRRM1 | 99 | 72.000 | ENSPANG00000035568 | - | 59 | 78.629 | Papio_anubis |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | ENSPANG00000010194 | SRRM1 | 100 | 98.901 | Papio_anubis |
| ENSG00000133226 | SRRM1 | 100 | 91.139 | ENSPMGG00000009036 | - | 73 | 92.308 | Periophthalmus_magnuspinnatus |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | ENSPEMG00000006254 | Srrm1 | 74 | 95.501 | Peromyscus_maniculatus_bairdii |
| ENSG00000133226 | SRRM1 | 100 | 94.937 | ENSPMEG00000002615 | - | 100 | 94.937 | Poecilia_mexicana |
| ENSG00000133226 | SRRM1 | 100 | 94.937 | ENSPMEG00000011776 | - | 100 | 94.937 | Poecilia_mexicana |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | ENSPPYG00000001716 | SRRM1 | 100 | 99.672 | Pongo_abelii |
| ENSG00000133226 | SRRM1 | 66 | 100.000 | ENSPCAG00000015138 | SRRM1 | 52 | 100.000 | Procavia_capensis |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | ENSPCOG00000021311 | SRRM1 | 100 | 98.468 | Propithecus_coquereli |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | ENSPVAG00000005836 | SRRM1 | 100 | 98.138 | Pteropus_vampyrus |
| ENSG00000133226 | SRRM1 | 99 | 94.872 | ENSPNYG00000005058 | - | 68 | 88.272 | Pundamilia_nyererei |
| ENSG00000133226 | SRRM1 | 99 | 93.590 | ENSPNAG00000005888 | - | 75 | 93.590 | Pygocentrus_nattereri |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | ENSRNOG00000018194 | Srrm1 | 75 | 95.646 | Rattus_norvegicus |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | ENSRBIG00000035461 | SRRM1 | 99 | 92.982 | Rhinopithecus_bieti |
| ENSG00000133226 | SRRM1 | 100 | 72.277 | ENSRROG00000041543 | - | 52 | 71.255 | Rhinopithecus_roxellana |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | ENSRROG00000035151 | SRRM1 | 100 | 99.671 | Rhinopithecus_roxellana |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | ENSSBOG00000019884 | SRRM1 | 100 | 100.000 | Saimiri_boliviensis_boliviensis |
| ENSG00000133226 | SRRM1 | 99 | 93.590 | ENSSMAG00000009318 | - | 58 | 92.647 | Scophthalmus_maximus |
| ENSG00000133226 | SRRM1 | 99 | 83.513 | ENSSARG00000013718 | SRRM1 | 91 | 83.513 | Sorex_araneus |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | ENSSPUG00000015890 | SRRM1 | 57 | 83.026 | Sphenodon_punctatus |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | ENSSSCG00000032947 | SRRM1 | 100 | 97.929 | Sus_scrofa |
| ENSG00000133226 | SRRM1 | 91 | 98.611 | ENSTGUG00000016669 | - | 99 | 100.000 | Taeniopygia_guttata |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | ENSTBEG00000000100 | SRRM1 | 100 | 97.700 | Tupaia_belangeri |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | ENSTTRG00000009365 | SRRM1 | 100 | 97.572 | Tursiops_truncatus |
| ENSG00000133226 | SRRM1 | 100 | 98.734 | ENSUAMG00000011740 | - | 87 | 100.000 | Ursus_americanus |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | ENSUMAG00000013121 | SRRM1 | 95 | 98.997 | Ursus_maritimus |
| ENSG00000133226 | SRRM1 | 100 | 100.000 | ENSVPAG00000007698 | SRRM1 | 100 | 93.764 | Vicugna_pacos |
| ENSG00000133226 | SRRM1 | 100 | 99.010 | ENSVVUG00000026966 | SRRM1 | 97 | 95.642 | Vulpes_vulpes |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000375 | RNA splicing, via transesterification reactions | 9531537. | TAS | Process |
| GO:0000398 | mRNA splicing, via spliceosome | 11991638. | IC | Process |
| GO:0000398 | mRNA splicing, via spliceosome | - | TAS | Process |
| GO:0003677 | DNA binding | - | IEA | Function |
| GO:0003723 | RNA binding | 22681889. | HDA | Function |
| GO:0005515 | protein binding | 25416956.26871637. | IPI | Function |
| GO:0005634 | nucleus | 9531537. | TAS | Component |
| GO:0005654 | nucleoplasm | - | TAS | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0006405 | RNA export from nucleus | - | TAS | Process |
| GO:0006406 | mRNA export from nucleus | - | TAS | Process |
| GO:0008380 | RNA splicing | 9531537. | TAS | Process |
| GO:0016363 | nuclear matrix | - | IEA | Component |
| GO:0016607 | nuclear speck | - | IDA | Component |
| GO:0031124 | mRNA 3'-end processing | - | TAS | Process |
| GO:0071013 | catalytic step 2 spliceosome | 11991638. | IDA | Component |