
| PID | Title | Article | Time | Author | Doi |
|---|---|---|---|---|---|
| 28984199 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000535570 | TAOK3-203 | 597 | - | ENSP00000443465 | 136 (aa) | - | F5H5E0 |
| ENST00000537822 | TAOK3-208 | 597 | - | ENSP00000439620 | 14 (aa) | - | F5H1I2 |
| ENST00000541186 | TAOK3-213 | 881 | - | ENSP00000438820 | 93 (aa) | - | F5H3L7 |
| ENST00000541878 | TAOK3-215 | 578 | - | ENSP00000444057 | 61 (aa) | - | F5H005 |
| ENST00000540561 | TAOK3-212 | 580 | - | ENSP00000443487 | 38 (aa) | - | F5H5C7 |
| ENST00000539872 | TAOK3-211 | 594 | - | ENSP00000438766 | 18 (aa) | - | F5H0M3 |
| ENST00000536584 | TAOK3-204 | 3229 | - | - | - (aa) | - | - |
| ENST00000537952 | TAOK3-209 | 2147 | - | ENSP00000443834 | 438 (aa) | - | G3V1Q8 |
| ENST00000538601 | TAOK3-210 | 555 | - | ENSP00000437389 | 185 (aa) | - | H0YF68 |
| ENST00000542902 | TAOK3-218 | 613 | - | ENSP00000440315 | 57 (aa) | - | F5GY38 |
| ENST00000541786 | TAOK3-214 | 590 | - | ENSP00000442541 | 21 (aa) | - | F5H7G4 |
| ENST00000542532 | TAOK3-216 | 641 | - | ENSP00000441071 | 36 (aa) | - | F5GX96 |
| ENST00000536979 | TAOK3-205 | 777 | - | ENSP00000441932 | 93 (aa) | - | F5GWV8 |
| ENST00000419821 | TAOK3-202 | 4301 | XM_006719445 | ENSP00000416374 | 898 (aa) | XP_006719508 | Q9H2K8 |
| ENST00000537569 | TAOK3-207 | 675 | - | - | - (aa) | - | - |
| ENST00000543709 | TAOK3-219 | 597 | - | - | - (aa) | - | - |
| ENST00000537305 | TAOK3-206 | 3932 | - | - | - (aa) | - | - |
| ENST00000542692 | TAOK3-217 | 588 | - | - | - (aa) | - | - |
| ENST00000392533 | TAOK3-201 | 4384 | XM_005253898 | ENSP00000376317 | 898 (aa) | XP_005253955 | Q9H2K8 |
| Pathway ID | Pathway Name | Source |
|---|---|---|
| hsa04010 | MAPK signaling pathway | KEGG |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000135090 | Glucose | 5E-6 | 25524916 |
| ENSG00000135090 | Insulin Resistance | 5E-6 | 25524916 |
| ENSG00000135090 | Glucose | 2E-6 | 25524916 |
| ENSG00000135090 | Insulin Resistance | 2E-6 | 25524916 |
| ENSG00000135090 | Bronchodilator Agents | 5E-6 | 26634245 |
| ENSG00000135090 | Forced Expiratory Volume | 5E-6 | 26634245 |
| ENSG00000135090 | Pulmonary Disease, Chronic Obstructive | 5E-6 | 26634245 |
| ENSG00000135090 | Bronchodilator Agents | 2E-6 | 26634245 |
| ENSG00000135090 | Forced Expiratory Volume | 2E-6 | 26634245 |
| ENSG00000135090 | Pulmonary Disease, Chronic Obstructive | 2E-6 | 26634245 |
| ENSG00000135090 | Morphine | 3E-7 | 24909733 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000135090 | rs650466 | 12 | 118208856 | T | Post bronchodilator FEV1 in COPD | 26634245 | NR unit decrease | 0.166 | EFO_0004314|GO_0097366|EFO_0000341 |
| ENSG00000135090 | rs61943529 | 12 | 118164051 | A | Post bronchodilator FEV1 in COPD | 26634245 | NR unit decrease | 0.164 | EFO_0004314|GO_0097366|EFO_0000341 |
| ENSG00000135090 | rs673078 | 12 | 118353636 | ? | Glucose homeostasis traits | 25524916 | [-0.0964-0.2564] unit decrease | 0.08 | EFO_0004471|EFO_0006896 |
| ENSG00000135090 | rs673078 | 12 | 118353636 | ? | Glucose homeostasis traits | 25524916 | [17.42-43.1] unit decrease | 30.26 | EFO_0004471|EFO_0006896 |
| ENSG00000135090 | rs795484 | 12 | 118152057 | A | Morphine dose requirement in tonsillectomy and adenoidectomy surgery | 24909733 | [10.0-21.1] unit increase | 16.1 | GO_0043278 |
| ENSG00000135090 | rs428073 | 12 | 118244946 | T | Red cell distribution width | 27863252 | [0.02-0.035] unit decrease | 0.02778707 | EFO_0005192 |
| ENSG00000135090 | rs2280711 | 12 | 118212149 | T | Neurociticism | 29500382 | z score increase | 6.29 | EFO_0007660 |
| ENSG00000135090 | rs6490177 | 12 | 118358280 | A | Neurociticism | 29500382 | z score increase | 6.54 | EFO_0007660 |
| ENSG00000135090 | rs6490177 | 12 | 118358280 | A | Neuroticism | 29942085 | z score increase | 6.046 | EFO_0007660 |
| ENSG00000135090 | rs353884 | 12 | 118225962 | ? | Red cell distribution width | 30595370 | EFO_0005192 | ||
| ENSG00000135090 | rs7314987 | 12 | 118274072 | T | Neuroticism | 30643256 | [0.0093-0.0181] unit increase | 0.013703822 | EFO_0007660 |
| ENSG00000135090 | rs7314987 | 12 | 118274072 | T | Depressive symptoms | 30643256 | [0.0058-0.0114] unit increase | 0.008617761 | EFO_0007006 |
| ENSG00000135090 | rs6490177 | 12 | 118358280 | ? | Neuroticism | 30595370 | EFO_0007660 | ||
| ENSG00000135090 | rs11068917 | 12 | 118353315 | A | Well-being spectrum (multivariate analysis) | 30643256 | [0.0052-0.0099] unit increase | 0.007531199 | EFO_0007869 |
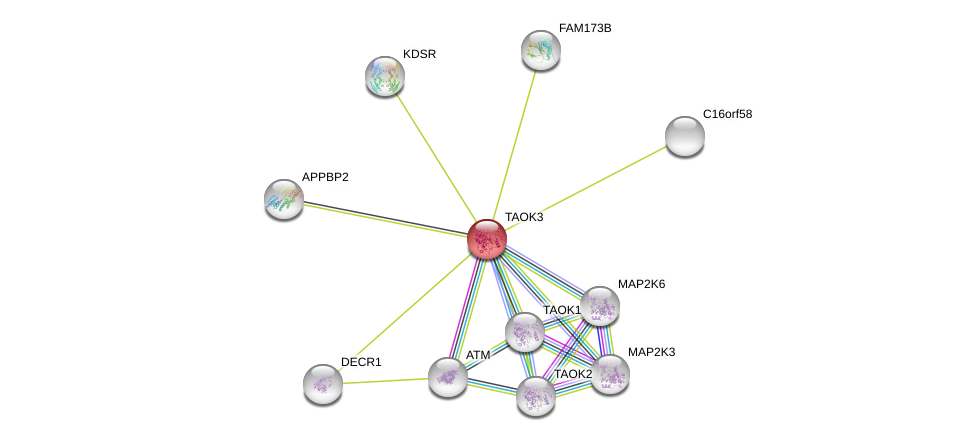
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000165 | MAPK cascade | 10924369. | IMP | Process |
| GO:0000186 | activation of MAPKK activity | 21873635. | IBA | Process |
| GO:0004674 | protein serine/threonine kinase activity | 21873635. | IBA | Function |
| GO:0004674 | protein serine/threonine kinase activity | 10559204. | IDA | Function |
| GO:0004709 | MAP kinase kinase kinase activity | 21873635. | IBA | Function |
| GO:0004860 | protein kinase inhibitor activity | 10559204. | TAS | Function |
| GO:0005515 | protein binding | 11278723. | IPI | Function |
| GO:0005524 | ATP binding | - | IEA | Function |
| GO:0005634 | nucleus | 21873635. | IBA | Component |
| GO:0005737 | cytoplasm | 21873635. | IBA | Component |
| GO:0005886 | plasma membrane | - | IEA | Component |
| GO:0006281 | DNA repair | - | IEA | Process |
| GO:0006468 | protein phosphorylation | 10559204. | IDA | Process |
| GO:0006468 | protein phosphorylation | 11278723. | IMP | Process |
| GO:0006469 | negative regulation of protein kinase activity | - | IEA | Process |
| GO:0006974 | cellular response to DNA damage stimulus | 17396146. | IDA | Process |
| GO:0007095 | mitotic G2 DNA damage checkpoint | 17396146. | IMP | Process |
| GO:0016740 | transferase activity | 10559204. | IDA | Function |
| GO:0023014 | signal transduction by protein phosphorylation | 21873635. | IBA | Process |
| GO:0031098 | stress-activated protein kinase signaling cascade | 21873635. | IBA | Process |
| GO:0032147 | activation of protein kinase activity | 21873635. | IBA | Process |
| GO:0032874 | positive regulation of stress-activated MAPK cascade | 17396146. | IMP | Process |
| GO:0043507 | positive regulation of JUN kinase activity | 11278723. | IMP | Process |
| GO:0046329 | negative regulation of JNK cascade | 10559204. | IDA | Process |
| GO:0046330 | positive regulation of JNK cascade | 10924369. | IMP | Process |
| GO:0046777 | protein autophosphorylation | 10559204. | IDA | Process |