
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000356667 | zf-CCCH | PF00642.24 | 0.00078 | 1 | 1 |
| ENSP00000258349 | zf-CCCH | PF00642.24 | 0.00078 | 1 | 1 |
| ENSP00000356669 | zf-CCCH | PF00642.24 | 0.00078 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000479099 | RC3H1-204 | 481 | - | - | - (aa) | - | - |
| ENST00000531594 | RC3H1-206 | 303 | - | - | - (aa) | - | - |
| ENST00000367694 | RC3H1-202 | 3775 | XM_011509231 | ENSP00000356667 | 1125 (aa) | XP_011507533 | Q5TC82 |
| ENST00000484867 | RC3H1-205 | 368 | - | - | - (aa) | - | - |
| ENST00000367696 | RC3H1-203 | 11261 | - | ENSP00000356669 | 1133 (aa) | - | Q5TC82 |
| ENST00000258349 | RC3H1-201 | 10988 | XM_005244921 | ENSP00000258349 | 1133 (aa) | XP_005244978 | Q5TC82 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000135870 | rs41304550 | 1 | 173937797 | T | Feeling miserable | 29500382 | z score decrease | 5.88 | EFO_0009598 |
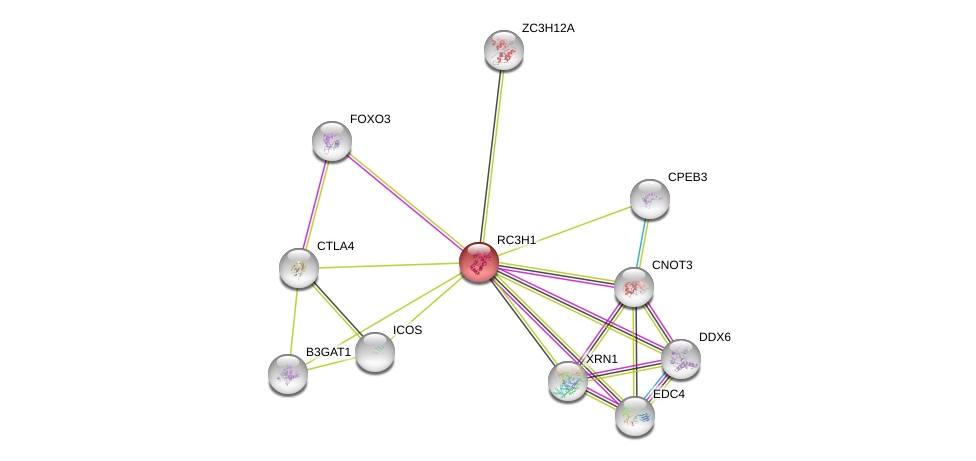
| Ensembl ID | Gene Symbol | Coverage | Identiy | Paralog | Gene Symbol | Coverage | Identiy |
|---|---|---|---|---|---|---|---|
| ENSG00000135870 | RC3H1 | 96 | 52.323 | ENSG00000056586 | RC3H2 | 94 | 83.223 |
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000135870 | RC3H1 | 100 | 96.911 | ENSAMEG00000015657 | RC3H1 | 100 | 96.911 | Ailuropoda_melanoleuca |
| ENSG00000135870 | RC3H1 | 100 | 88.056 | ENSAPLG00000005341 | RC3H1 | 100 | 88.056 | Anas_platyrhynchos |
| ENSG00000135870 | RC3H1 | 100 | 85.253 | ENSACAG00000015271 | RC3H1 | 100 | 85.253 | Anolis_carolinensis |
| ENSG00000135870 | RC3H1 | 100 | 98.941 | ENSANAG00000036716 | RC3H1 | 100 | 98.941 | Aotus_nancymaae |
| ENSG00000135870 | RC3H1 | 100 | 96.533 | ENSBTAG00000052708 | RC3H1 | 100 | 96.533 | Bos_taurus |
| ENSG00000135870 | RC3H1 | 100 | 98.588 | ENSCJAG00000009106 | RC3H1 | 100 | 98.588 | Callithrix_jacchus |
| ENSG00000135870 | RC3H1 | 100 | 96.999 | ENSCAFG00000014434 | RC3H1 | 100 | 96.999 | Canis_familiaris |
| ENSG00000135870 | RC3H1 | 100 | 96.914 | ENSCAFG00020017112 | RC3H1 | 99 | 98.202 | Canis_lupus_dingo |
| ENSG00000135870 | RC3H1 | 100 | 96.889 | ENSCHIG00000002873 | RC3H1 | 100 | 96.889 | Capra_hircus |
| ENSG00000135870 | RC3H1 | 99 | 95.322 | ENSTSYG00000003913 | RC3H1 | 99 | 95.322 | Carlito_syrichta |
| ENSG00000135870 | RC3H1 | 100 | 91.534 | ENSCPOG00000004349 | RC3H1 | 100 | 91.446 | Cavia_porcellus |
| ENSG00000135870 | RC3H1 | 100 | 96.558 | ENSCCAG00000025652 | RC3H1 | 100 | 96.558 | Cebus_capucinus |
| ENSG00000135870 | RC3H1 | 100 | 99.294 | ENSCATG00000033497 | RC3H1 | 100 | 99.294 | Cercocebus_atys |
| ENSG00000135870 | RC3H1 | 100 | 94.974 | ENSCLAG00000011267 | RC3H1 | 100 | 94.974 | Chinchilla_lanigera |
| ENSG00000135870 | RC3H1 | 96 | 99.079 | ENSCSAG00000011823 | RC3H1 | 85 | 99.079 | Chlorocebus_sabaeus |
| ENSG00000135870 | RC3H1 | 100 | 89.737 | ENSCPBG00000014670 | RC3H1 | 100 | 89.737 | Chrysemys_picta_bellii |
| ENSG00000135870 | RC3H1 | 100 | 99.295 | ENSCANG00000032120 | RC3H1 | 100 | 99.295 | Colobus_angolensis_palliatus |
| ENSG00000135870 | RC3H1 | 100 | 94.263 | ENSCGRG00001020812 | Rc3h1 | 100 | 93.822 | Cricetulus_griseus_chok1gshd |
| ENSG00000135870 | RC3H1 | 100 | 94.263 | ENSCGRG00000011952 | Rc3h1 | 100 | 93.822 | Cricetulus_griseus_crigri |
| ENSG00000135870 | RC3H1 | 55 | 95.484 | ENSDNOG00000011494 | RC3H1 | 99 | 95.484 | Dasypus_novemcinctus |
| ENSG00000135870 | RC3H1 | 100 | 96.028 | ENSDORG00000004855 | Rc3h1 | 100 | 96.028 | Dipodomys_ordii |
| ENSG00000135870 | RC3H1 | 82 | 96.527 | ENSETEG00000012064 | RC3H1 | 87 | 96.527 | Echinops_telfairi |
| ENSG00000135870 | RC3H1 | 100 | 96.622 | ENSEASG00005017000 | RC3H1 | 100 | 96.622 | Equus_asinus_asinus |
| ENSG00000135870 | RC3H1 | 100 | 96.444 | ENSECAG00000021208 | RC3H1 | 100 | 96.444 | Equus_caballus |
| ENSG00000135870 | RC3H1 | 83 | 97.059 | ENSEEUG00000014861 | RC3H1 | 93 | 83.302 | Erinaceus_europaeus |
| ENSG00000135870 | RC3H1 | 100 | 97.352 | ENSFCAG00000014924 | RC3H1 | 100 | 97.352 | Felis_catus |
| ENSG00000135870 | RC3H1 | 100 | 87.424 | ENSFALG00000002947 | RC3H1 | 100 | 86.900 | Ficedula_albicollis |
| ENSG00000135870 | RC3H1 | 100 | 94.974 | ENSFDAG00000013303 | RC3H1 | 100 | 94.974 | Fukomys_damarensis |
| ENSG00000135870 | RC3H1 | 100 | 87.898 | ENSGALG00000004587 | RC3H1 | 100 | 87.898 | Gallus_gallus |
| ENSG00000135870 | RC3H1 | 100 | 88.703 | ENSGAGG00000018194 | RC3H1 | 100 | 88.967 | Gopherus_agassizii |
| ENSG00000135870 | RC3H1 | 100 | 92.876 | ENSGGOG00000005735 | RC3H1 | 100 | 92.876 | Gorilla_gorilla |
| ENSG00000135870 | RC3H1 | 88 | 88.846 | ENSHGLG00000004183 | - | 99 | 88.846 | Heterocephalus_glaber_female |
| ENSG00000135870 | RC3H1 | 100 | 94.444 | ENSHGLG00100008577 | RC3H1 | 100 | 94.444 | Heterocephalus_glaber_male |
| ENSG00000135870 | RC3H1 | 100 | 96.649 | ENSSTOG00000016033 | RC3H1 | 100 | 96.649 | Ictidomys_tridecemlineatus |
| ENSG00000135870 | RC3H1 | 100 | 95.322 | ENSJJAG00000018915 | Rc3h1 | 100 | 95.146 | Jaculus_jaculus |
| ENSG00000135870 | RC3H1 | 100 | 75.217 | ENSLACG00000016186 | RC3H1 | 100 | 78.279 | Latimeria_chalumnae |
| ENSG00000135870 | RC3H1 | 97 | 97.541 | ENSLAFG00000004367 | RC3H1 | 100 | 97.541 | Loxodonta_africana |
| ENSG00000135870 | RC3H1 | 100 | 99.206 | ENSMFAG00000033725 | RC3H1 | 100 | 99.206 | Macaca_fascicularis |
| ENSG00000135870 | RC3H1 | 100 | 99.117 | ENSMMUG00000000630 | RC3H1 | 100 | 99.117 | Macaca_mulatta |
| ENSG00000135870 | RC3H1 | 100 | 99.294 | ENSMNEG00000035968 | RC3H1 | 100 | 99.294 | Macaca_nemestrina |
| ENSG00000135870 | RC3H1 | 100 | 99.206 | ENSMLEG00000037455 | RC3H1 | 100 | 99.206 | Mandrillus_leucophaeus |
| ENSG00000135870 | RC3H1 | 97 | 94.254 | ENSMAUG00000020864 | Rc3h1 | 100 | 93.750 | Mesocricetus_auratus |
| ENSG00000135870 | RC3H1 | 100 | 97.440 | ENSMICG00000003000 | RC3H1 | 100 | 97.440 | Microcebus_murinus |
| ENSG00000135870 | RC3H1 | 93 | 92.898 | ENSMOCG00000016587 | Rc3h1 | 98 | 92.519 | Microtus_ochrogaster |
| ENSG00000135870 | RC3H1 | 100 | 92.240 | ENSMODG00000004289 | RC3H1 | 100 | 92.328 | Monodelphis_domestica |
| ENSG00000135870 | RC3H1 | 100 | 93.822 | MGP_CAROLIEiJ_G0014756 | Rc3h1 | 100 | 93.380 | Mus_caroli |
| ENSG00000135870 | RC3H1 | 100 | 93.645 | ENSMUSG00000040423 | Rc3h1 | 100 | 93.204 | Mus_musculus |
| ENSG00000135870 | RC3H1 | 100 | 93.469 | MGP_PahariEiJ_G0027992 | Rc3h1 | 100 | 93.027 | Mus_pahari |
| ENSG00000135870 | RC3H1 | 100 | 93.469 | MGP_SPRETEiJ_G0015563 | Rc3h1 | 100 | 93.027 | Mus_spretus |
| ENSG00000135870 | RC3H1 | 100 | 96.649 | ENSMPUG00000004747 | RC3H1 | 100 | 96.649 | Mustela_putorius_furo |
| ENSG00000135870 | RC3H1 | 100 | 95.587 | ENSMLUG00000011127 | RC3H1 | 100 | 95.587 | Myotis_lucifugus |
| ENSG00000135870 | RC3H1 | 100 | 95.852 | ENSNGAG00000011135 | Rc3h1 | 100 | 95.410 | Nannospalax_galili |
| ENSG00000135870 | RC3H1 | 100 | 99.382 | ENSNLEG00000015327 | RC3H1 | 100 | 99.382 | Nomascus_leucogenys |
| ENSG00000135870 | RC3H1 | 99 | 82.935 | ENSOPRG00000013068 | RC3H1 | 99 | 83.024 | Ochotona_princeps |
| ENSG00000135870 | RC3H1 | 100 | 94.533 | ENSODEG00000014185 | RC3H1 | 100 | 94.533 | Octodon_degus |
| ENSG00000135870 | RC3H1 | 100 | 91.441 | ENSOANG00000000924 | RC3H1 | 100 | 91.354 | Ornithorhynchus_anatinus |
| ENSG00000135870 | RC3H1 | 99 | 96.150 | ENSOCUG00000004930 | RC3H1 | 98 | 96.150 | Oryctolagus_cuniculus |
| ENSG00000135870 | RC3H1 | 100 | 90.220 | ENSOGAG00000004044 | RC3H1 | 100 | 90.220 | Otolemur_garnettii |
| ENSG00000135870 | RC3H1 | 100 | 96.491 | ENSOARG00000013011 | RC3H1 | 100 | 96.491 | Ovis_aries |
| ENSG00000135870 | RC3H1 | 100 | 99.824 | ENSPPAG00000039253 | RC3H1 | 100 | 99.824 | Pan_paniscus |
| ENSG00000135870 | RC3H1 | 100 | 97.333 | ENSPPRG00000009007 | RC3H1 | 100 | 97.333 | Panthera_pardus |
| ENSG00000135870 | RC3H1 | 100 | 97.012 | ENSPTIG00000015326 | RC3H1 | 100 | 97.012 | Panthera_tigris_altaica |
| ENSG00000135870 | RC3H1 | 100 | 99.824 | ENSPTRG00000001702 | RC3H1 | 100 | 99.824 | Pan_troglodytes |
| ENSG00000135870 | RC3H1 | 100 | 99.206 | ENSPANG00000008886 | RC3H1 | 100 | 99.206 | Papio_anubis |
| ENSG00000135870 | RC3H1 | 100 | 89.497 | ENSPSIG00000016181 | RC3H1 | 100 | 89.762 | Pelodiscus_sinensis |
| ENSG00000135870 | RC3H1 | 100 | 93.822 | ENSPEMG00000011966 | Rc3h1 | 100 | 93.469 | Peromyscus_maniculatus_bairdii |
| ENSG00000135870 | RC3H1 | 70 | 94.051 | ENSPCIG00000002058 | RC3H1 | 95 | 98.430 | Phascolarctos_cinereus |
| ENSG00000135870 | RC3H1 | 100 | 96.028 | ENSPPYG00000000490 | RC3H1 | 98 | 99.421 | Pongo_abelii |
| ENSG00000135870 | RC3H1 | 81 | 100.000 | ENSPCAG00000010775 | RC3H1 | 87 | 100.000 | Procavia_capensis |
| ENSG00000135870 | RC3H1 | 83 | 96.902 | ENSPCOG00000021795 | RC3H1 | 96 | 96.902 | Propithecus_coquereli |
| ENSG00000135870 | RC3H1 | 100 | 88.173 | ENSPVAG00000007593 | RC3H1 | 100 | 88.173 | Pteropus_vampyrus |
| ENSG00000135870 | RC3H1 | 97 | 62.422 | ENSPNAG00000023439 | rc3h1a | 95 | 90.991 | Pygocentrus_nattereri |
| ENSG00000135870 | RC3H1 | 100 | 99.206 | ENSRBIG00000037743 | RC3H1 | 100 | 99.206 | Rhinopithecus_bieti |
| ENSG00000135870 | RC3H1 | 100 | 99.117 | ENSRROG00000036546 | RC3H1 | 100 | 99.117 | Rhinopithecus_roxellana |
| ENSG00000135870 | RC3H1 | 93 | 93.702 | ENSSBOG00000010664 | RC3H1 | 100 | 93.702 | Saimiri_boliviensis_boliviensis |
| ENSG00000135870 | RC3H1 | 100 | 91.454 | ENSSHAG00000007589 | RC3H1 | 100 | 91.542 | Sarcophilus_harrisii |
| ENSG00000135870 | RC3H1 | 87 | 86.712 | ENSSARG00000004660 | RC3H1 | 86 | 86.712 | Sorex_araneus |
| ENSG00000135870 | RC3H1 | 100 | 84.501 | ENSSPUG00000018472 | RC3H1 | 100 | 83.961 | Sphenodon_punctatus |
| ENSG00000135870 | RC3H1 | 100 | 97.529 | ENSSSCG00000015498 | RC3H1 | 100 | 97.529 | Sus_scrofa |
| ENSG00000135870 | RC3H1 | 100 | 87.895 | ENSTGUG00000004090 | RC3H1 | 100 | 87.895 | Taeniopygia_guttata |
| ENSG00000135870 | RC3H1 | 89 | 97.561 | ENSTBEG00000005015 | RC3H1 | 89 | 97.561 | Tupaia_belangeri |
| ENSG00000135870 | RC3H1 | 100 | 96.564 | ENSTTRG00000008572 | RC3H1 | 100 | 96.564 | Tursiops_truncatus |
| ENSG00000135870 | RC3H1 | 100 | 96.293 | ENSUAMG00000019525 | RC3H1 | 100 | 96.293 | Ursus_americanus |
| ENSG00000135870 | RC3H1 | 100 | 96.825 | ENSUMAG00000020087 | RC3H1 | 100 | 96.825 | Ursus_maritimus |
| ENSG00000135870 | RC3H1 | 91 | 98.649 | ENSVPAG00000002591 | RC3H1 | 92 | 98.649 | Vicugna_pacos |
| ENSG00000135870 | RC3H1 | 100 | 96.658 | ENSVVUG00000014201 | RC3H1 | 100 | 96.658 | Vulpes_vulpes |
| ENSG00000135870 | RC3H1 | 100 | 79.199 | ENSXETG00000020030 | rc3h1 | 100 | 78.746 | Xenopus_tropicalis |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000209 | protein polyubiquitination | 26489670. | IDA | Process |
| GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | - | ISS | Process |
| GO:0000932 | P-body | - | ISS | Component |
| GO:0000956 | nuclear-transcribed mRNA catabolic process | 25026078. | IDA | Process |
| GO:0000956 | nuclear-transcribed mRNA catabolic process | - | ISS | Process |
| GO:0001782 | B cell homeostasis | - | IEA | Process |
| GO:0002634 | regulation of germinal center formation | - | ISS | Process |
| GO:0002635 | negative regulation of germinal center formation | - | ISS | Process |
| GO:0003723 | RNA binding | 22658674.22681889. | HDA | Function |
| GO:0003725 | double-stranded RNA binding | 25026078.25504471. | IDA | Function |
| GO:0003730 | mRNA 3'-UTR binding | - | ISS | Function |
| GO:0004842 | ubiquitin-protein transferase activity | 26489670. | IDA | Function |
| GO:0005515 | protein binding | 25697406. | IPI | Function |
| GO:0008270 | zinc ion binding | - | ISS | Function |
| GO:0010494 | cytoplasmic stress granule | - | ISS | Component |
| GO:0010608 | posttranscriptional regulation of gene expression | - | ISS | Process |
| GO:0030889 | negative regulation of B cell proliferation | - | ISS | Process |
| GO:0033962 | cytoplasmic mRNA processing body assembly | - | ISS | Process |
| GO:0035198 | miRNA binding | - | ISS | Function |
| GO:0035613 | RNA stem-loop binding | 25504471. | IDA | Function |
| GO:0035613 | RNA stem-loop binding | - | ISS | Function |
| GO:0042098 | T cell proliferation | - | IEA | Process |
| GO:0043029 | T cell homeostasis | - | IEA | Process |
| GO:0043488 | regulation of mRNA stability | - | ISS | Process |
| GO:0045623 | negative regulation of T-helper cell differentiation | - | ISS | Process |
| GO:0046007 | negative regulation of activated T cell proliferation | - | ISS | Process |
| GO:0048535 | lymph node development | - | IEA | Process |
| GO:0048536 | spleen development | - | IEA | Process |
| GO:0050852 | T cell receptor signaling pathway | - | ISS | Process |
| GO:0050856 | regulation of T cell receptor signaling pathway | - | ISS | Process |
| GO:0061014 | positive regulation of mRNA catabolic process | - | ISS | Process |
| GO:0061158 | 3'-UTR-mediated mRNA destabilization | - | ISS | Process |
| GO:0061470 | T follicular helper cell differentiation | - | IEA | Process |
| GO:0061630 | ubiquitin protein ligase activity | 19217324. | TAS | Function |
| GO:0071347 | cellular response to interleukin-1 | - | ISS | Process |
| GO:1901224 | positive regulation of NIK/NF-kappaB signaling | - | IEA | Process |
| GO:2000320 | negative regulation of T-helper 17 cell differentiation | - | ISS | Process |
| GO:2000628 | regulation of miRNA metabolic process | - | ISS | Process |