
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000384617 | W2 | PF02020.18 | 1.7e-20 | 1 | 1 |
| ENSP00000385577 | W2 | PF02020.18 | 3.7e-20 | 1 | 1 |
| ENSP00000258761 | W2 | PF02020.18 | 5.1e-20 | 1 | 1 |
| ENSP00000397249 | W2 | PF02020.18 | 5.1e-20 | 1 | 1 |
| ENSP00000403481 | W2 | PF02020.18 | 6.9e-16 | 1 | 1 |
| ENSP00000411715 | W2 | PF02020.18 | 4e-15 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000430000 | BZW2-205 | 617 | - | ENSP00000416531 | 162 (aa) | - | C9JF98 |
| ENST00000415365 | BZW2-204 | 1405 | - | ENSP00000403481 | 408 (aa) | - | E7ETZ4 |
| ENST00000630952 | BZW2-214 | 1721 | - | ENSP00000486454 | 161 (aa) | - | E9PFE3 |
| ENST00000446596 | BZW2-211 | 859 | - | ENSP00000412750 | 274 (aa) | - | Q75MG1 |
| ENST00000433922 | BZW2-207 | 1862 | - | ENSP00000397249 | 419 (aa) | - | Q9Y6E2 |
| ENST00000436868 | BZW2-208 | 1784 | - | ENSP00000389183 | 170 (aa) | - | F8WDX8 |
| ENST00000480517 | BZW2-213 | 2101 | - | - | - (aa) | - | - |
| ENST00000432311 | BZW2-206 | 800 | - | - | - (aa) | - | - |
| ENST00000258761 | BZW2-201 | 1860 | XM_006715706 | ENSP00000258761 | 419 (aa) | XP_006715769 | Q9Y6E2 |
| ENST00000438834 | BZW2-210 | 674 | - | ENSP00000415924 | 208 (aa) | - | E7EMS9 |
| ENST00000405202 | BZW2-202 | 1689 | - | ENSP00000385577 | 343 (aa) | - | B5MCH7 |
| ENST00000407633 | BZW2-203 | 898 | XM_024446735 | ENSP00000384617 | 225 (aa) | XP_024302503 | B5MCE7 |
| ENST00000437745 | BZW2-209 | 1711 | - | ENSP00000406395 | 161 (aa) | - | E9PFE3 |
| ENST00000452975 | BZW2-212 | 1464 | - | ENSP00000411715 | 189 (aa) | - | E9PFD4 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000136261 | rs706027 | 7 | 16660352 | T | High light scatter reticulocyte percentage of red cells | 27863252 | [0.033-0.048] unit increase | 0.04043556 | EFO_0007986 |
| ENSG00000136261 | rs696281 | 7 | 16691417 | T | High light scatter reticulocyte percentage of red cells | 27863252 | [0.041-0.058] unit decrease | 0.04915672 | EFO_0007986 |
| ENSG00000136261 | rs706027 | 7 | 16660352 | T | Reticulocyte count | 27863252 | [0.03-0.045] unit increase | 0.03734367 | EFO_0007986 |
| ENSG00000136261 | rs696281 | 7 | 16691417 | T | Reticulocyte count | 27863252 | [0.037-0.055] unit decrease | 0.04597321 | EFO_0007986 |
| ENSG00000136261 | rs696279 | 7 | 16661173 | A | Mean platelet volume | 27863252 | [0.015-0.029] unit decrease | 0.02204623 | EFO_0004584 |
| ENSG00000136261 | rs706042 | 7 | 16696157 | ? | Velopharyngeal dysfunction | 29855589 | [NR] unit decrease | 0.12076525 | EFO_0009336 |
| ENSG00000136261 | rs58370486 | 7 | 16668236 | ? | Alzheimer's disease (cognitive decline) | 23535033 | unit decrease | 0.36 | EFO_0000249 |
| ENSG00000136261 | rs696281 | 7 | 16691417 | T | Reticulocyte fraction of red cells | 27863252 | [0.039-0.057] unit decrease | 0.04801943 | EFO_0007986 |
| ENSG00000136261 | rs706027 | 7 | 16660352 | T | Reticulocyte fraction of red cells | 27863252 | [0.032-0.046] unit increase | 0.03900041 | EFO_0007986 |
| ENSG00000136261 | rs706027 | 7 | 16660352 | T | Immature fraction of reticulocytes | 27863252 | [0.023-0.037] unit increase | 0.03022379 | EFO_0007986 |
| ENSG00000136261 | rs706027 | 7 | 16660352 | T | High light scatter reticulocyte count | 27863252 | [0.032-0.046] unit increase | 0.03911746 | EFO_0007986 |
| ENSG00000136261 | rs696281 | 7 | 16691417 | T | High light scatter reticulocyte count | 27863252 | [0.039-0.056] unit decrease | 0.04782177 | EFO_0007986 |
| ENSG00000136261 | rs116196735 | 7 | 16670980 | ? | Diastolic blood pressure x smoking status (current vs non-current) interaction (2df test) | 29455858 | EFO_0006527|EFO_0006336 | ||
| ENSG00000136261 | rs116196735 | 7 | 16670980 | ? | Systolic blood pressure x smoking status (current vs non-current) interaction (2df test) | 29455858 | EFO_0006527|EFO_0006335 | ||
| ENSG00000136261 | rs706027 | 7 | 16660352 | ? | Red cell distribution width | 30595370 | EFO_0005192 | ||
| ENSG00000136261 | rs706027 | 7 | 16660352 | ? | Mean corpuscular hemoglobin | 30595370 | EFO_0004527 |
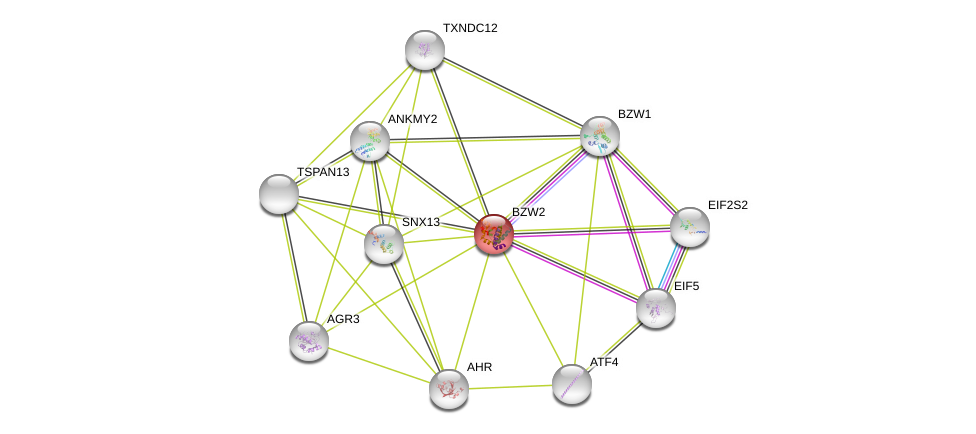
| Ensembl ID | Gene Symbol | Coverage | Identiy | Paralog | Gene Symbol | Coverage | Identiy |
|---|---|---|---|---|---|---|---|
| ENSG00000136261 | BZW2 | 99 | 72.099 | ENSG00000082153 | BZW1 | 98 | 71.875 |
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000136261 | BZW2 | 99 | 70.443 | ENSAPOG00000011525 | - | 91 | 70.443 | Acanthochromis_polyacanthus |
| ENSG00000136261 | BZW2 | 99 | 71.429 | ENSAPOG00000019670 | bzw1a | 96 | 71.182 | Acanthochromis_polyacanthus |
| ENSG00000136261 | BZW2 | 99 | 71.852 | ENSAMEG00000005859 | BZW1 | 89 | 71.921 | Ailuropoda_melanoleuca |
| ENSG00000136261 | BZW2 | 100 | 100.000 | ENSAMEG00000010473 | BZW2 | 99 | 99.284 | Ailuropoda_melanoleuca |
| ENSG00000136261 | BZW2 | 99 | 71.921 | ENSACIG00000024418 | bzw1a | 96 | 71.921 | Amphilophus_citrinellus |
| ENSG00000136261 | BZW2 | 99 | 70.625 | ENSACIG00000013845 | - | 96 | 69.951 | Amphilophus_citrinellus |
| ENSG00000136261 | BZW2 | 99 | 71.875 | ENSAOCG00000005035 | bzw1a | 96 | 71.429 | Amphiprion_ocellaris |
| ENSG00000136261 | BZW2 | 99 | 70.443 | ENSAOCG00000017269 | - | 96 | 70.443 | Amphiprion_ocellaris |
| ENSG00000136261 | BZW2 | 99 | 71.429 | ENSAPEG00000003772 | bzw1a | 96 | 71.182 | Amphiprion_percula |
| ENSG00000136261 | BZW2 | 99 | 70.443 | ENSAPEG00000010528 | - | 96 | 70.443 | Amphiprion_percula |
| ENSG00000136261 | BZW2 | 99 | 70.807 | ENSATEG00000004179 | - | 96 | 70.443 | Anabas_testudineus |
| ENSG00000136261 | BZW2 | 99 | 71.675 | ENSATEG00000013087 | bzw1a | 95 | 71.675 | Anabas_testudineus |
| ENSG00000136261 | BZW2 | 100 | 96.444 | ENSAPLG00000008006 | BZW2 | 97 | 94.118 | Anas_platyrhynchos |
| ENSG00000136261 | BZW2 | 99 | 72.500 | ENSAPLG00000016194 | BZW1 | 96 | 71.394 | Anas_platyrhynchos |
| ENSG00000136261 | BZW2 | 99 | 73.125 | ENSACAG00000016934 | BZW1 | 81 | 72.906 | Anolis_carolinensis |
| ENSG00000136261 | BZW2 | 100 | 91.932 | ENSACAG00000012646 | BZW2 | 97 | 91.932 | Anolis_carolinensis |
| ENSG00000136261 | BZW2 | 99 | 72.099 | ENSANAG00000021489 | BZW1 | 93 | 70.690 | Aotus_nancymaae |
| ENSG00000136261 | BZW2 | 100 | 100.000 | ENSANAG00000004675 | BZW2 | 100 | 100.000 | Aotus_nancymaae |
| ENSG00000136261 | BZW2 | 100 | 71.605 | ENSACLG00000014272 | bzw1a | 96 | 71.182 | Astatotilapia_calliptera |
| ENSG00000136261 | BZW2 | 99 | 70.625 | ENSACLG00000008855 | - | 95 | 69.704 | Astatotilapia_calliptera |
| ENSG00000136261 | BZW2 | 99 | 83.292 | ENSAMXG00000011012 | bzw2 | 97 | 83.292 | Astyanax_mexicanus |
| ENSG00000136261 | BZW2 | 99 | 72.050 | ENSAMXG00000016810 | bzw1b | 96 | 70.936 | Astyanax_mexicanus |
| ENSG00000136261 | BZW2 | 99 | 71.875 | ENSAMXG00000002926 | bzw1a | 95 | 71.182 | Astyanax_mexicanus |
| ENSG00000136261 | BZW2 | 99 | 72.099 | ENSBTAG00000006049 | BZW1 | 96 | 72.167 | Bos_taurus |
| ENSG00000136261 | BZW2 | 100 | 100.000 | ENSBTAG00000014262 | BZW2 | 82 | 99.284 | Bos_taurus |
| ENSG00000136261 | BZW2 | 99 | 72.167 | ENSCJAG00000004267 | BZW1 | 89 | 72.167 | Callithrix_jacchus |
| ENSG00000136261 | BZW2 | 100 | 100.000 | ENSCJAG00000000954 | BZW2 | 100 | 100.000 | Callithrix_jacchus |
| ENSG00000136261 | BZW2 | 99 | 72.099 | ENSCAFG00000011878 | BZW1 | 89 | 72.167 | Canis_familiaris |
| ENSG00000136261 | BZW2 | 100 | 100.000 | ENSCAFG00000002424 | BZW2 | 100 | 100.000 | Canis_familiaris |
| ENSG00000136261 | BZW2 | 93 | 68.095 | ENSCAFG00020021462 | - | 93 | 68.246 | Canis_lupus_dingo |
| ENSG00000136261 | BZW2 | 99 | 72.099 | ENSCAFG00020004433 | - | 89 | 72.167 | Canis_lupus_dingo |
| ENSG00000136261 | BZW2 | 100 | 100.000 | ENSCAFG00020001541 | BZW2 | 100 | 100.000 | Canis_lupus_dingo |
| ENSG00000136261 | BZW2 | 100 | 99.635 | ENSCHIG00000020691 | BZW2 | 100 | 99.523 | Capra_hircus |
| ENSG00000136261 | BZW2 | 99 | 72.099 | ENSCHIG00000019964 | BZW1 | 96 | 72.167 | Capra_hircus |
| ENSG00000136261 | BZW2 | 100 | 100.000 | ENSTSYG00000009806 | BZW2 | 100 | 99.523 | Carlito_syrichta |
| ENSG00000136261 | BZW2 | 98 | 70.440 | ENSTSYG00000029674 | - | 93 | 58.519 | Carlito_syrichta |
| ENSG00000136261 | BZW2 | 99 | 72.099 | ENSTSYG00000013638 | - | 96 | 72.167 | Carlito_syrichta |
| ENSG00000136261 | BZW2 | 91 | 100.000 | ENSCAPG00000003500 | BZW2 | 97 | 84.804 | Cavia_aperea |
| ENSG00000136261 | BZW2 | 97 | 71.299 | ENSCAPG00000015172 | BZW1 | 94 | 70.000 | Cavia_aperea |
| ENSG00000136261 | BZW2 | 99 | 51.485 | ENSCPOG00000023007 | - | 92 | 51.485 | Cavia_porcellus |
| ENSG00000136261 | BZW2 | 99 | 71.875 | ENSCPOG00000010449 | - | 96 | 71.921 | Cavia_porcellus |
| ENSG00000136261 | BZW2 | 100 | 99.556 | ENSCPOG00000011515 | BZW2 | 97 | 88.480 | Cavia_porcellus |
| ENSG00000136261 | BZW2 | 100 | 100.000 | ENSCCAG00000028749 | BZW2 | 100 | 100.000 | Cebus_capucinus |
| ENSG00000136261 | BZW2 | 99 | 72.167 | ENSCCAG00000012616 | BZW1 | 96 | 72.167 | Cebus_capucinus |
| ENSG00000136261 | BZW2 | 99 | 72.167 | ENSCATG00000032512 | BZW1 | 96 | 72.167 | Cercocebus_atys |
| ENSG00000136261 | BZW2 | 100 | 100.000 | ENSCATG00000040929 | BZW2 | 100 | 100.000 | Cercocebus_atys |
| ENSG00000136261 | BZW2 | 100 | 100.000 | ENSCLAG00000011809 | BZW2 | 100 | 100.000 | Chinchilla_lanigera |
| ENSG00000136261 | BZW2 | 99 | 71.875 | ENSCLAG00000012079 | BZW1 | 96 | 71.921 | Chinchilla_lanigera |
| ENSG00000136261 | BZW2 | 99 | 72.167 | ENSCSAG00000010957 | BZW1 | 96 | 72.167 | Chlorocebus_sabaeus |
| ENSG00000136261 | BZW2 | 100 | 100.000 | ENSCSAG00000013474 | BZW2 | 100 | 100.000 | Chlorocebus_sabaeus |
| ENSG00000136261 | BZW2 | 83 | 96.923 | ENSCHOG00000011030 | BZW2 | 100 | 80.392 | Choloepus_hoffmanni |
| ENSG00000136261 | BZW2 | 99 | 71.875 | ENSCHOG00000003543 | - | 95 | 58.867 | Choloepus_hoffmanni |
| ENSG00000136261 | BZW2 | 99 | 72.500 | ENSCPBG00000011755 | BZW1 | 96 | 72.167 | Chrysemys_picta_bellii |
| ENSG00000136261 | BZW2 | 100 | 96.889 | ENSCPBG00000021873 | BZW2 | 95 | 94.363 | Chrysemys_picta_bellii |
| ENSG00000136261 | BZW2 | 99 | 72.167 | ENSCANG00000041550 | BZW1 | 96 | 72.167 | Colobus_angolensis_palliatus |
| ENSG00000136261 | BZW2 | 100 | 100.000 | ENSCANG00000008829 | BZW2 | 99 | 99.041 | Colobus_angolensis_palliatus |
| ENSG00000136261 | BZW2 | 98 | 82.390 | ENSCGRG00001021112 | - | 97 | 76.961 | Cricetulus_griseus_chok1gshd |
| ENSG00000136261 | BZW2 | 100 | 99.383 | ENSCGRG00001013113 | - | 100 | 98.807 | Cricetulus_griseus_chok1gshd |
| ENSG00000136261 | BZW2 | 99 | 72.500 | ENSCGRG00001017178 | Bzw1 | 96 | 72.414 | Cricetulus_griseus_chok1gshd |
| ENSG00000136261 | BZW2 | 99 | 83.230 | ENSCGRG00000009430 | - | 97 | 78.398 | Cricetulus_griseus_crigri |
| ENSG00000136261 | BZW2 | 99 | 72.500 | ENSCGRG00000002341 | Bzw1 | 95 | 72.414 | Cricetulus_griseus_crigri |
| ENSG00000136261 | BZW2 | 100 | 99.383 | ENSCGRG00000009735 | - | 100 | 98.807 | Cricetulus_griseus_crigri |
| ENSG00000136261 | BZW2 | 99 | 70.625 | ENSCSEG00000003418 | bzw1a | 96 | 68.719 | Cynoglossus_semilaevis |
| ENSG00000136261 | BZW2 | 97 | 65.861 | ENSCSEG00000019691 | - | 93 | 66.067 | Cynoglossus_semilaevis |
| ENSG00000136261 | BZW2 | 99 | 69.375 | ENSCVAG00000001975 | - | 96 | 67.734 | Cyprinodon_variegatus |
| ENSG00000136261 | BZW2 | 99 | 71.429 | ENSCVAG00000022879 | bzw1a | 96 | 70.690 | Cyprinodon_variegatus |
| ENSG00000136261 | BZW2 | 99 | 85.625 | ENSDARG00000035918 | bzw2 | 97 | 84.275 | Danio_rerio |
| ENSG00000136261 | BZW2 | 99 | 71.250 | ENSDARG00000010481 | bzw1a | 96 | 70.443 | Danio_rerio |
| ENSG00000136261 | BZW2 | 99 | 72.050 | ENSDARG00000099148 | bzw1b | 96 | 69.458 | Danio_rerio |
| ENSG00000136261 | BZW2 | 100 | 100.000 | ENSDNOG00000001663 | BZW2 | 83 | 99.284 | Dasypus_novemcinctus |
| ENSG00000136261 | BZW2 | 99 | 71.358 | ENSDNOG00000035002 | - | 95 | 71.429 | Dasypus_novemcinctus |
| ENSG00000136261 | BZW2 | 99 | 72.099 | ENSDORG00000029865 | Bzw1 | 96 | 72.167 | Dipodomys_ordii |
| ENSG00000136261 | BZW2 | 100 | 100.000 | ENSDORG00000010878 | Bzw2 | 100 | 99.761 | Dipodomys_ordii |
| ENSG00000136261 | BZW2 | 99 | 55.901 | FBgn0250753 | kra | 96 | 49.507 | Drosophila_melanogaster |
| ENSG00000136261 | BZW2 | 94 | 73.460 | ENSETEG00000008565 | BZW1 | 97 | 69.588 | Echinops_telfairi |
| ENSG00000136261 | BZW2 | 90 | 99.010 | ENSETEG00000000863 | BZW2 | 74 | 100.000 | Echinops_telfairi |
| ENSG00000136261 | BZW2 | 99 | 70.552 | ENSEBUG00000008484 | bzw1a | 84 | 68.215 | Eptatretus_burgeri |
| ENSG00000136261 | BZW2 | 99 | 72.167 | ENSEASG00005000922 | BZW1 | 96 | 72.167 | Equus_asinus_asinus |
| ENSG00000136261 | BZW2 | 100 | 100.000 | ENSEASG00005021624 | BZW2 | 100 | 99.761 | Equus_asinus_asinus |
| ENSG00000136261 | BZW2 | 100 | 100.000 | ENSECAG00000022038 | BZW2 | 100 | 99.761 | Equus_caballus |
| ENSG00000136261 | BZW2 | 99 | 72.099 | ENSECAG00000011768 | BZW1 | 96 | 72.167 | Equus_caballus |
| ENSG00000136261 | BZW2 | 97 | 71.601 | ENSEEUG00000008717 | BZW1 | 97 | 71.649 | Erinaceus_europaeus |
| ENSG00000136261 | BZW2 | 100 | 99.556 | ENSEEUG00000001289 | BZW2 | 100 | 88.305 | Erinaceus_europaeus |
| ENSG00000136261 | BZW2 | 98 | 70.886 | ENSELUG00000008110 | - | 96 | 68.812 | Esox_lucius |
| ENSG00000136261 | BZW2 | 99 | 71.250 | ENSELUG00000015556 | bzw1a | 96 | 68.227 | Esox_lucius |
| ENSG00000136261 | BZW2 | 99 | 83.125 | ENSELUG00000023531 | bzw2 | 97 | 80.835 | Esox_lucius |
| ENSG00000136261 | BZW2 | 99 | 72.099 | ENSFCAG00000025654 | BZW1 | 95 | 72.167 | Felis_catus |
| ENSG00000136261 | BZW2 | 100 | 100.000 | ENSFCAG00000009109 | BZW2 | 100 | 100.000 | Felis_catus |
| ENSG00000136261 | BZW2 | 99 | 73.125 | ENSFALG00000004161 | BZW1 | 95 | 72.167 | Ficedula_albicollis |
| ENSG00000136261 | BZW2 | 100 | 96.444 | ENSFALG00000009931 | BZW2 | 97 | 94.118 | Ficedula_albicollis |
| ENSG00000136261 | BZW2 | 100 | 100.000 | ENSFDAG00000014900 | BZW2 | 100 | 99.761 | Fukomys_damarensis |
| ENSG00000136261 | BZW2 | 99 | 71.875 | ENSFDAG00000013117 | BZW1 | 93 | 71.675 | Fukomys_damarensis |
| ENSG00000136261 | BZW2 | 84 | 73.684 | ENSFHEG00000013200 | bzw1a | 86 | 68.203 | Fundulus_heteroclitus |
| ENSG00000136261 | BZW2 | 99 | 68.323 | ENSFHEG00000006742 | - | 96 | 67.734 | Fundulus_heteroclitus |
| ENSG00000136261 | BZW2 | 99 | 70.807 | ENSGMOG00000001477 | BZW1 | 96 | 68.473 | Gadus_morhua |
| ENSG00000136261 | BZW2 | 97 | 67.069 | ENSGMOG00000003853 | - | 97 | 67.784 | Gadus_morhua |
| ENSG00000136261 | BZW2 | 100 | 94.118 | ENSGALG00000010809 | BZW2 | 99 | 94.118 | Gallus_gallus |
| ENSG00000136261 | BZW2 | 99 | 72.500 | ENSGALG00000008220 | BZW1 | 97 | 71.883 | Gallus_gallus |
| ENSG00000136261 | BZW2 | 99 | 68.944 | ENSGAFG00000016335 | - | 91 | 68.227 | Gambusia_affinis |
| ENSG00000136261 | BZW2 | 99 | 68.116 | ENSGAFG00000000253 | bzw1a | 96 | 68.193 | Gambusia_affinis |
| ENSG00000136261 | BZW2 | 99 | 73.750 | ENSGACG00000015646 | bzw1a | 96 | 70.690 | Gasterosteus_aculeatus |
| ENSG00000136261 | BZW2 | 99 | 70.186 | ENSGACG00000007556 | - | 96 | 69.951 | Gasterosteus_aculeatus |
| ENSG00000136261 | BZW2 | 99 | 72.500 | ENSGAGG00000010463 | BZW1 | 98 | 72.167 | Gopherus_agassizii |
| ENSG00000136261 | BZW2 | 94 | 85.475 | ENSGAGG00000015424 | - | 93 | 91.667 | Gopherus_agassizii |
| ENSG00000136261 | BZW2 | 99 | 72.099 | ENSGGOG00000016296 | BZW1 | 96 | 72.167 | Gorilla_gorilla |
| ENSG00000136261 | BZW2 | 100 | 100.000 | ENSGGOG00000022223 | BZW2 | 100 | 100.000 | Gorilla_gorilla |
| ENSG00000136261 | BZW2 | 99 | 70.625 | ENSHBUG00000018698 | - | 89 | 69.704 | Haplochromis_burtoni |
| ENSG00000136261 | BZW2 | 99 | 71.875 | ENSHBUG00000009482 | bzw1a | 96 | 71.182 | Haplochromis_burtoni |
| ENSG00000136261 | BZW2 | 100 | 100.000 | ENSHGLG00000011038 | BZW2 | 100 | 99.761 | Heterocephalus_glaber_female |
| ENSG00000136261 | BZW2 | 99 | 71.875 | ENSHGLG00000011961 | BZW1 | 95 | 71.921 | Heterocephalus_glaber_female |
| ENSG00000136261 | BZW2 | 100 | 100.000 | ENSHGLG00100011287 | BZW2 | 100 | 99.284 | Heterocephalus_glaber_male |
| ENSG00000136261 | BZW2 | 99 | 71.852 | ENSHGLG00100003983 | - | 84 | 71.921 | Heterocephalus_glaber_male |
| ENSG00000136261 | BZW2 | 99 | 71.875 | ENSHCOG00000008122 | bzw1a | 90 | 87.500 | Hippocampus_comes |
| ENSG00000136261 | BZW2 | 99 | 71.875 | ENSIPUG00000003696 | bzw1a | 96 | 70.197 | Ictalurus_punctatus |
| ENSG00000136261 | BZW2 | 99 | 85.625 | ENSIPUG00000021662 | bzw2 | 97 | 85.258 | Ictalurus_punctatus |
| ENSG00000136261 | BZW2 | 100 | 100.000 | ENSSTOG00000002506 | BZW2 | 100 | 99.761 | Ictidomys_tridecemlineatus |
| ENSG00000136261 | BZW2 | 97 | 63.746 | ENSSTOG00000030689 | - | 96 | 65.097 | Ictidomys_tridecemlineatus |
| ENSG00000136261 | BZW2 | 99 | 69.533 | ENSSTOG00000031155 | - | 96 | 69.608 | Ictidomys_tridecemlineatus |
| ENSG00000136261 | BZW2 | 100 | 100.000 | ENSJJAG00000016668 | Bzw2 | 100 | 99.761 | Jaculus_jaculus |
| ENSG00000136261 | BZW2 | 99 | 72.099 | ENSJJAG00000022065 | Bzw1 | 89 | 72.167 | Jaculus_jaculus |
| ENSG00000136261 | BZW2 | 99 | 71.875 | ENSKMAG00000003931 | bzw1a | 83 | 71.429 | Kryptolebias_marmoratus |
| ENSG00000136261 | BZW2 | 99 | 68.750 | ENSKMAG00000016408 | - | 96 | 67.241 | Kryptolebias_marmoratus |
| ENSG00000136261 | BZW2 | 99 | 70.625 | ENSLBEG00000021455 | - | 96 | 69.951 | Labrus_bergylta |
| ENSG00000136261 | BZW2 | 99 | 72.500 | ENSLBEG00000010918 | bzw1a | 96 | 70.197 | Labrus_bergylta |
| ENSG00000136261 | BZW2 | 100 | 89.951 | ENSLACG00000007218 | BZW2 | 84 | 89.951 | Latimeria_chalumnae |
| ENSG00000136261 | BZW2 | 99 | 71.921 | ENSLACG00000002968 | BZW1 | 96 | 71.921 | Latimeria_chalumnae |
| ENSG00000136261 | BZW2 | 99 | 72.500 | ENSLOCG00000010532 | bzw1a | 95 | 71.675 | Lepisosteus_oculatus |
| ENSG00000136261 | BZW2 | 93 | 83.529 | ENSLOCG00000011315 | bzw2 | 96 | 82.199 | Lepisosteus_oculatus |
| ENSG00000136261 | BZW2 | 100 | 99.519 | ENSLAFG00000003942 | BZW2 | 100 | 99.284 | Loxodonta_africana |
| ENSG00000136261 | BZW2 | 99 | 72.099 | ENSLAFG00000009883 | BZW1 | 95 | 72.167 | Loxodonta_africana |
| ENSG00000136261 | BZW2 | 100 | 100.000 | ENSMFAG00000002705 | BZW2 | 100 | 95.238 | Macaca_fascicularis |
| ENSG00000136261 | BZW2 | 99 | 72.167 | ENSMFAG00000020809 | BZW1 | 95 | 72.167 | Macaca_fascicularis |
| ENSG00000136261 | BZW2 | 100 | 100.000 | ENSMMUG00000017045 | BZW2 | 100 | 100.000 | Macaca_mulatta |
| ENSG00000136261 | BZW2 | 99 | 72.167 | ENSMMUG00000022202 | BZW1 | 95 | 72.167 | Macaca_mulatta |
| ENSG00000136261 | BZW2 | 99 | 72.167 | ENSMNEG00000043638 | BZW1 | 96 | 72.167 | Macaca_nemestrina |
| ENSG00000136261 | BZW2 | 100 | 100.000 | ENSMNEG00000002080 | BZW2 | 100 | 100.000 | Macaca_nemestrina |
| ENSG00000136261 | BZW2 | 100 | 100.000 | ENSMLEG00000033305 | BZW2 | 100 | 94.792 | Mandrillus_leucophaeus |
| ENSG00000136261 | BZW2 | 99 | 80.165 | ENSMLEG00000018894 | BZW1 | 88 | 80.165 | Mandrillus_leucophaeus |
| ENSG00000136261 | BZW2 | 99 | 71.429 | ENSMAMG00000022490 | bzw1a | 96 | 71.429 | Mastacembelus_armatus |
| ENSG00000136261 | BZW2 | 99 | 69.458 | ENSMAMG00000014694 | - | 96 | 69.458 | Mastacembelus_armatus |
| ENSG00000136261 | BZW2 | 100 | 71.605 | ENSMZEG00005015758 | bzw1a | 96 | 71.182 | Maylandia_zebra |
| ENSG00000136261 | BZW2 | 99 | 70.625 | ENSMZEG00005002198 | - | 96 | 69.704 | Maylandia_zebra |
| ENSG00000136261 | BZW2 | 97 | 71.299 | ENSMGAG00000007562 | BZW1 | 94 | 70.603 | Meleagris_gallopavo |
| ENSG00000136261 | BZW2 | 100 | 94.118 | ENSMGAG00000010129 | BZW2 | 97 | 94.118 | Meleagris_gallopavo |
| ENSG00000136261 | BZW2 | 99 | 71.429 | ENSMAUG00000004813 | Bzw1 | 89 | 69.458 | Mesocricetus_auratus |
| ENSG00000136261 | BZW2 | 100 | 99.270 | ENSMAUG00000000123 | Bzw2 | 100 | 98.807 | Mesocricetus_auratus |
| ENSG00000136261 | BZW2 | 99 | 72.167 | ENSMICG00000003355 | BZW1 | 96 | 72.167 | Microcebus_murinus |
| ENSG00000136261 | BZW2 | 100 | 100.000 | ENSMICG00000015293 | BZW2 | 100 | 99.761 | Microcebus_murinus |
| ENSG00000136261 | BZW2 | 99 | 72.500 | ENSMOCG00000008375 | Bzw1 | 96 | 72.414 | Microtus_ochrogaster |
| ENSG00000136261 | BZW2 | 100 | 99.635 | ENSMOCG00000020705 | Bzw2 | 100 | 99.284 | Microtus_ochrogaster |
| ENSG00000136261 | BZW2 | 99 | 71.875 | ENSMMOG00000012820 | bzw1a | 95 | 69.531 | Mola_mola |
| ENSG00000136261 | BZW2 | 99 | 68.750 | ENSMMOG00000005563 | - | 96 | 64.039 | Mola_mola |
| ENSG00000136261 | BZW2 | 97 | 94.578 | ENSMODG00000001582 | BZW2 | 91 | 94.859 | Monodelphis_domestica |
| ENSG00000136261 | BZW2 | 99 | 72.099 | ENSMODG00000015000 | BZW1 | 96 | 72.167 | Monodelphis_domestica |
| ENSG00000136261 | BZW2 | 94 | 76.364 | ENSMALG00000018187 | - | 96 | 67.726 | Monopterus_albus |
| ENSG00000136261 | BZW2 | 99 | 71.429 | ENSMALG00000006867 | bzw1a | 96 | 71.429 | Monopterus_albus |
| ENSG00000136261 | BZW2 | 99 | 72.099 | MGP_CAROLIEiJ_G0014162 | Bzw1 | 96 | 72.167 | Mus_caroli |
| ENSG00000136261 | BZW2 | 100 | 99.635 | MGP_CAROLIEiJ_G0017704 | Bzw2 | 100 | 99.045 | Mus_caroli |
| ENSG00000136261 | BZW2 | 99 | 72.099 | ENSMUSG00000051223 | Bzw1 | 96 | 72.167 | Mus_musculus |
| ENSG00000136261 | BZW2 | 100 | 99.635 | ENSMUSG00000020547 | Bzw2 | 100 | 99.045 | Mus_musculus |
| ENSG00000136261 | BZW2 | 99 | 71.429 | MGP_PahariEiJ_G0027400 | Bzw1 | 96 | 63.547 | Mus_pahari |
| ENSG00000136261 | BZW2 | 100 | 99.038 | MGP_PahariEiJ_G0029462 | Bzw2 | 91 | 98.568 | Mus_pahari |
| ENSG00000136261 | BZW2 | 99 | 72.167 | MGP_SPRETEiJ_G0014968 | Bzw1 | 96 | 72.167 | Mus_spretus |
| ENSG00000136261 | BZW2 | 100 | 99.635 | MGP_SPRETEiJ_G0018547 | Bzw2 | 100 | 99.045 | Mus_spretus |
| ENSG00000136261 | BZW2 | 100 | 100.000 | ENSMPUG00000014004 | BZW2 | 100 | 100.000 | Mustela_putorius_furo |
| ENSG00000136261 | BZW2 | 99 | 71.921 | ENSMPUG00000012344 | BZW1 | 95 | 71.990 | Mustela_putorius_furo |
| ENSG00000136261 | BZW2 | 99 | 72.167 | ENSMLUG00000001653 | BZW1 | 96 | 72.236 | Myotis_lucifugus |
| ENSG00000136261 | BZW2 | 99 | 99.554 | ENSMLUG00000014943 | BZW2 | 99 | 97.857 | Myotis_lucifugus |
| ENSG00000136261 | BZW2 | 100 | 99.635 | ENSNGAG00000015192 | Bzw2 | 100 | 99.284 | Nannospalax_galili |
| ENSG00000136261 | BZW2 | 99 | 72.099 | ENSNGAG00000014200 | Bzw1 | 96 | 72.167 | Nannospalax_galili |
| ENSG00000136261 | BZW2 | 99 | 70.625 | ENSNBRG00000012864 | - | 96 | 64.778 | Neolamprologus_brichardi |
| ENSG00000136261 | BZW2 | 99 | 71.429 | ENSNBRG00000015102 | bzw1a | 96 | 71.182 | Neolamprologus_brichardi |
| ENSG00000136261 | BZW2 | 100 | 100.000 | ENSNLEG00000016417 | BZW2 | 100 | 100.000 | Nomascus_leucogenys |
| ENSG00000136261 | BZW2 | 99 | 72.099 | ENSNLEG00000006929 | BZW1 | 96 | 72.167 | Nomascus_leucogenys |
| ENSG00000136261 | BZW2 | 94 | 62.434 | ENSMEUG00000009847 | - | 54 | 86.842 | Notamacropus_eugenii |
| ENSG00000136261 | BZW2 | 95 | 94.393 | ENSMEUG00000000858 | BZW2 | 97 | 87.990 | Notamacropus_eugenii |
| ENSG00000136261 | BZW2 | 100 | 98.667 | ENSOPRG00000002795 | - | 100 | 97.887 | Ochotona_princeps |
| ENSG00000136261 | BZW2 | 99 | 71.250 | ENSOPRG00000005652 | BZW1 | 95 | 58.374 | Ochotona_princeps |
| ENSG00000136261 | BZW2 | 100 | 100.000 | ENSOPRG00000005256 | - | 100 | 98.568 | Ochotona_princeps |
| ENSG00000136261 | BZW2 | 100 | 99.556 | ENSODEG00000007505 | BZW2 | 100 | 99.523 | Octodon_degus |
| ENSG00000136261 | BZW2 | 95 | 76.596 | ENSODEG00000008411 | - | 98 | 65.271 | Octodon_degus |
| ENSG00000136261 | BZW2 | 99 | 70.625 | ENSONIG00000004328 | - | 96 | 69.704 | Oreochromis_niloticus |
| ENSG00000136261 | BZW2 | 99 | 71.429 | ENSONIG00000012258 | bzw1a | 96 | 71.182 | Oreochromis_niloticus |
| ENSG00000136261 | BZW2 | 88 | 58.497 | ENSOANG00000010824 | - | 96 | 58.333 | Ornithorhynchus_anatinus |
| ENSG00000136261 | BZW2 | 61 | 97.080 | ENSOANG00000020501 | - | 100 | 97.080 | Ornithorhynchus_anatinus |
| ENSG00000136261 | BZW2 | 99 | 71.921 | ENSOCUG00000010647 | BZW1 | 95 | 71.990 | Oryctolagus_cuniculus |
| ENSG00000136261 | BZW2 | 100 | 100.000 | ENSOCUG00000016419 | BZW2 | 99 | 99.518 | Oryctolagus_cuniculus |
| ENSG00000136261 | BZW2 | 99 | 71.875 | ENSORLG00000000117 | bzw1a | 96 | 70.443 | Oryzias_latipes |
| ENSG00000136261 | BZW2 | 99 | 70.625 | ENSORLG00000018257 | - | 96 | 69.212 | Oryzias_latipes |
| ENSG00000136261 | BZW2 | 99 | 70.186 | ENSORLG00020009773 | - | 96 | 64.286 | Oryzias_latipes_hni |
| ENSG00000136261 | BZW2 | 99 | 71.875 | ENSORLG00020011814 | bzw1a | 96 | 70.443 | Oryzias_latipes_hni |
| ENSG00000136261 | BZW2 | 99 | 71.875 | ENSORLG00015014130 | bzw1a | 96 | 70.690 | Oryzias_latipes_hsok |
| ENSG00000136261 | BZW2 | 99 | 70.186 | ENSORLG00015006345 | - | 88 | 68.719 | Oryzias_latipes_hsok |
| ENSG00000136261 | BZW2 | 100 | 71.111 | ENSOMEG00000019988 | bzw1a | 96 | 71.675 | Oryzias_melastigma |
| ENSG00000136261 | BZW2 | 99 | 70.186 | ENSOMEG00000014106 | - | 98 | 68.719 | Oryzias_melastigma |
| ENSG00000136261 | BZW2 | 100 | 100.000 | ENSOGAG00000007600 | BZW2 | 100 | 99.761 | Otolemur_garnettii |
| ENSG00000136261 | BZW2 | 99 | 72.099 | ENSOGAG00000012326 | BZW1 | 89 | 72.167 | Otolemur_garnettii |
| ENSG00000136261 | BZW2 | 100 | 99.519 | ENSOARG00000008855 | BZW2 | 96 | 98.807 | Ovis_aries |
| ENSG00000136261 | BZW2 | 99 | 71.921 | ENSOARG00000016520 | BZW1 | 89 | 71.990 | Ovis_aries |
| ENSG00000136261 | BZW2 | 100 | 100.000 | ENSPPAG00000029762 | BZW2 | 100 | 100.000 | Pan_paniscus |
| ENSG00000136261 | BZW2 | 99 | 72.099 | ENSPPAG00000027971 | BZW1 | 96 | 72.167 | Pan_paniscus |
| ENSG00000136261 | BZW2 | 100 | 100.000 | ENSPPRG00000023909 | BZW2 | 100 | 100.000 | Panthera_pardus |
| ENSG00000136261 | BZW2 | 99 | 72.099 | ENSPPRG00000021144 | BZW1 | 96 | 72.167 | Panthera_pardus |
| ENSG00000136261 | BZW2 | 99 | 72.099 | ENSPTIG00000014068 | BZW1 | 96 | 72.167 | Panthera_tigris_altaica |
| ENSG00000136261 | BZW2 | 100 | 100.000 | ENSPTIG00000013171 | BZW2 | 96 | 97.176 | Panthera_tigris_altaica |
| ENSG00000136261 | BZW2 | 100 | 100.000 | ENSPTRG00000018951 | BZW2 | 100 | 100.000 | Pan_troglodytes |
| ENSG00000136261 | BZW2 | 99 | 72.099 | ENSPTRG00000012791 | BZW1 | 96 | 72.167 | Pan_troglodytes |
| ENSG00000136261 | BZW2 | 99 | 72.167 | ENSPANG00000016459 | BZW1 | 95 | 72.167 | Papio_anubis |
| ENSG00000136261 | BZW2 | 100 | 100.000 | ENSPANG00000025373 | BZW2 | 100 | 100.000 | Papio_anubis |
| ENSG00000136261 | BZW2 | 99 | 71.429 | ENSPKIG00000012285 | - | 96 | 70.690 | Paramormyrops_kingsleyae |
| ENSG00000136261 | BZW2 | 99 | 71.875 | ENSPKIG00000020584 | bzw1a | 96 | 70.690 | Paramormyrops_kingsleyae |
| ENSG00000136261 | BZW2 | 99 | 85.625 | ENSPKIG00000010815 | bzw2 | 92 | 85.258 | Paramormyrops_kingsleyae |
| ENSG00000136261 | BZW2 | 100 | 91.975 | ENSPSIG00000008748 | BZW2 | 97 | 91.443 | Pelodiscus_sinensis |
| ENSG00000136261 | BZW2 | 99 | 72.500 | ENSPSIG00000003613 | BZW1 | 98 | 71.173 | Pelodiscus_sinensis |
| ENSG00000136261 | BZW2 | 99 | 71.875 | ENSPMGG00000004836 | bzw1a | 95 | 62.315 | Periophthalmus_magnuspinnatus |
| ENSG00000136261 | BZW2 | 97 | 68.580 | ENSPMGG00000004823 | - | 96 | 68.580 | Periophthalmus_magnuspinnatus |
| ENSG00000136261 | BZW2 | 78 | 85.227 | ENSPMGG00000020531 | BZW2 | 94 | 85.227 | Periophthalmus_magnuspinnatus |
| ENSG00000136261 | BZW2 | 99 | 72.099 | ENSPEMG00000013940 | - | 96 | 72.167 | Peromyscus_maniculatus_bairdii |
| ENSG00000136261 | BZW2 | 100 | 99.111 | ENSPEMG00000019563 | Bzw2 | 100 | 98.807 | Peromyscus_maniculatus_bairdii |
| ENSG00000136261 | BZW2 | 99 | 71.703 | ENSPMAG00000000412 | - | 96 | 71.703 | Petromyzon_marinus |
| ENSG00000136261 | BZW2 | 99 | 71.638 | ENSPMAG00000004075 | - | 97 | 71.638 | Petromyzon_marinus |
| ENSG00000136261 | BZW2 | 100 | 95.255 | ENSPCIG00000008586 | BZW2 | 97 | 94.118 | Phascolarctos_cinereus |
| ENSG00000136261 | BZW2 | 99 | 72.167 | ENSPCIG00000019483 | - | 95 | 72.167 | Phascolarctos_cinereus |
| ENSG00000136261 | BZW2 | 99 | 71.429 | ENSPFOG00000004908 | bzw1a | 96 | 70.936 | Poecilia_formosa |
| ENSG00000136261 | BZW2 | 99 | 68.944 | ENSPFOG00000003280 | - | 96 | 67.805 | Poecilia_formosa |
| ENSG00000136261 | BZW2 | 99 | 68.944 | ENSPLAG00000016928 | - | 96 | 68.473 | Poecilia_latipinna |
| ENSG00000136261 | BZW2 | 99 | 71.429 | ENSPLAG00000002665 | bzw1a | 96 | 71.182 | Poecilia_latipinna |
| ENSG00000136261 | BZW2 | 99 | 71.429 | ENSPMEG00000009549 | bzw1a | 96 | 70.936 | Poecilia_mexicana |
| ENSG00000136261 | BZW2 | 99 | 68.944 | ENSPMEG00000018902 | - | 96 | 68.227 | Poecilia_mexicana |
| ENSG00000136261 | BZW2 | 99 | 68.944 | ENSPREG00000015136 | - | 96 | 68.473 | Poecilia_reticulata |
| ENSG00000136261 | BZW2 | 99 | 71.429 | ENSPREG00000021850 | bzw1a | 93 | 85.417 | Poecilia_reticulata |
| ENSG00000136261 | BZW2 | 100 | 100.000 | ENSPPYG00000017763 | BZW2 | 100 | 100.000 | Pongo_abelii |
| ENSG00000136261 | BZW2 | 97 | 68.262 | ENSPPYG00000013060 | BZW1 | 97 | 68.342 | Pongo_abelii |
| ENSG00000136261 | BZW2 | 100 | 99.111 | ENSPCAG00000001577 | BZW2 | 83 | 99.160 | Procavia_capensis |
| ENSG00000136261 | BZW2 | 100 | 99.556 | ENSPCOG00000014275 | BZW2 | 97 | 88.480 | Propithecus_coquereli |
| ENSG00000136261 | BZW2 | 99 | 60.248 | ENSPCOG00000016767 | - | 94 | 51.597 | Propithecus_coquereli |
| ENSG00000136261 | BZW2 | 99 | 72.167 | ENSPCOG00000015831 | - | 96 | 72.167 | Propithecus_coquereli |
| ENSG00000136261 | BZW2 | 100 | 100.000 | ENSPVAG00000014571 | BZW2 | 100 | 99.761 | Pteropus_vampyrus |
| ENSG00000136261 | BZW2 | 94 | 66.667 | ENSPVAG00000008111 | BZW1 | 96 | 57.732 | Pteropus_vampyrus |
| ENSG00000136261 | BZW2 | 99 | 71.875 | ENSPNYG00000022122 | bzw1a | 96 | 71.182 | Pundamilia_nyererei |
| ENSG00000136261 | BZW2 | 99 | 70.625 | ENSPNYG00000006245 | - | 89 | 69.704 | Pundamilia_nyererei |
| ENSG00000136261 | BZW2 | 99 | 85.625 | ENSPNAG00000009810 | bzw2 | 97 | 84.767 | Pygocentrus_nattereri |
| ENSG00000136261 | BZW2 | 99 | 71.250 | ENSPNAG00000007226 | bzw1a | 96 | 70.443 | Pygocentrus_nattereri |
| ENSG00000136261 | BZW2 | 99 | 71.429 | ENSPNAG00000018846 | bzw1b | 89 | 70.936 | Pygocentrus_nattereri |
| ENSG00000136261 | BZW2 | 99 | 72.099 | ENSRNOG00000013977 | Bzw1 | 96 | 72.167 | Rattus_norvegicus |
| ENSG00000136261 | BZW2 | 100 | 99.635 | ENSRNOG00000005096 | Bzw2 | 100 | 98.807 | Rattus_norvegicus |
| ENSG00000136261 | BZW2 | 100 | 100.000 | ENSRBIG00000031981 | BZW2 | 100 | 100.000 | Rhinopithecus_bieti |
| ENSG00000136261 | BZW2 | 99 | 71.429 | ENSRBIG00000031336 | BZW1 | 95 | 71.788 | Rhinopithecus_bieti |
| ENSG00000136261 | BZW2 | 99 | 72.167 | ENSRROG00000044305 | BZW1 | 96 | 72.167 | Rhinopithecus_roxellana |
| ENSG00000136261 | BZW2 | 100 | 100.000 | ENSRROG00000038313 | BZW2 | 100 | 100.000 | Rhinopithecus_roxellana |
| ENSG00000136261 | BZW2 | 100 | 100.000 | ENSSBOG00000023271 | BZW2 | 100 | 100.000 | Saimiri_boliviensis_boliviensis |
| ENSG00000136261 | BZW2 | 99 | 72.099 | ENSSBOG00000020710 | BZW1 | 96 | 72.167 | Saimiri_boliviensis_boliviensis |
| ENSG00000136261 | BZW2 | 99 | 72.099 | ENSSHAG00000012368 | BZW1 | 95 | 72.167 | Sarcophilus_harrisii |
| ENSG00000136261 | BZW2 | 100 | 95.255 | ENSSHAG00000004361 | BZW2 | 95 | 94.853 | Sarcophilus_harrisii |
| ENSG00000136261 | BZW2 | 99 | 85.625 | ENSSFOG00015015434 | bzw2 | 72 | 84.521 | Scleropages_formosus |
| ENSG00000136261 | BZW2 | 99 | 71.875 | ENSSFOG00015019024 | bzw1a | 96 | 70.936 | Scleropages_formosus |
| ENSG00000136261 | BZW2 | 99 | 71.250 | ENSSFOG00015007027 | BZW1 | 96 | 70.690 | Scleropages_formosus |
| ENSG00000136261 | BZW2 | 99 | 71.875 | ENSSMAG00000013262 | bzw1a | 96 | 70.443 | Scophthalmus_maximus |
| ENSG00000136261 | BZW2 | 99 | 71.250 | ENSSMAG00000005715 | - | 96 | 64.286 | Scophthalmus_maximus |
| ENSG00000136261 | BZW2 | 99 | 70.000 | ENSSDUG00000011658 | - | 96 | 69.951 | Seriola_dumerili |
| ENSG00000136261 | BZW2 | 99 | 84.375 | ENSSDUG00000009414 | bzw2 | 97 | 83.784 | Seriola_dumerili |
| ENSG00000136261 | BZW2 | 99 | 67.734 | ENSSDUG00000013408 | bzw1a | 96 | 67.813 | Seriola_dumerili |
| ENSG00000136261 | BZW2 | 98 | 71.053 | ENSSLDG00000000310 | BZW2 | 84 | 87.179 | Seriola_lalandi_dorsalis |
| ENSG00000136261 | BZW2 | 99 | 70.000 | ENSSLDG00000022189 | - | 96 | 69.951 | Seriola_lalandi_dorsalis |
| ENSG00000136261 | BZW2 | 86 | 83.420 | ENSSLDG00000011675 | bzw2 | 84 | 85.405 | Seriola_lalandi_dorsalis |
| ENSG00000136261 | BZW2 | 99 | 71.429 | ENSSLDG00000010836 | bzw1a | 96 | 71.182 | Seriola_lalandi_dorsalis |
| ENSG00000136261 | BZW2 | 97 | 71.601 | ENSSARG00000009734 | BZW1 | 97 | 71.649 | Sorex_araneus |
| ENSG00000136261 | BZW2 | 100 | 98.765 | ENSSARG00000002955 | BZW2 | 97 | 92.402 | Sorex_araneus |
| ENSG00000136261 | BZW2 | 99 | 72.500 | ENSSPUG00000003041 | BZW1 | 96 | 72.414 | Sphenodon_punctatus |
| ENSG00000136261 | BZW2 | 100 | 93.627 | ENSSPUG00000011687 | BZW2 | 94 | 93.627 | Sphenodon_punctatus |
| ENSG00000136261 | BZW2 | 99 | 71.875 | ENSSPAG00000002996 | bzw1a | 96 | 71.429 | Stegastes_partitus |
| ENSG00000136261 | BZW2 | 99 | 70.690 | ENSSPAG00000015804 | - | 96 | 70.690 | Stegastes_partitus |
| ENSG00000136261 | BZW2 | 99 | 72.099 | ENSSSCG00000016094 | BZW1 | 96 | 72.167 | Sus_scrofa |
| ENSG00000136261 | BZW2 | 100 | 100.000 | ENSSSCG00000023118 | BZW2 | 100 | 100.000 | Sus_scrofa |
| ENSG00000136261 | BZW2 | 100 | 96.444 | ENSTGUG00000002567 | - | 97 | 94.118 | Taeniopygia_guttata |
| ENSG00000136261 | BZW2 | 99 | 73.125 | ENSTGUG00000010398 | BZW1 | 95 | 72.167 | Taeniopygia_guttata |
| ENSG00000136261 | BZW2 | 99 | 71.875 | ENSTRUG00000005902 | bzw1a | 96 | 69.951 | Takifugu_rubripes |
| ENSG00000136261 | BZW2 | 99 | 70.186 | ENSTRUG00000005576 | - | 95 | 68.719 | Takifugu_rubripes |
| ENSG00000136261 | BZW2 | 99 | 69.091 | ENSTNIG00000017044 | - | 95 | 67.961 | Tetraodon_nigroviridis |
| ENSG00000136261 | BZW2 | 99 | 71.875 | ENSTNIG00000017173 | bzw1a | 96 | 70.443 | Tetraodon_nigroviridis |
| ENSG00000136261 | BZW2 | 97 | 65.257 | ENSTBEG00000000430 | BZW1 | 97 | 66.237 | Tupaia_belangeri |
| ENSG00000136261 | BZW2 | 99 | 71.875 | ENSTTRG00000003639 | BZW1 | 95 | 58.867 | Tursiops_truncatus |
| ENSG00000136261 | BZW2 | 100 | 99.556 | ENSTTRG00000009867 | BZW2 | 100 | 98.807 | Tursiops_truncatus |
| ENSG00000136261 | BZW2 | 99 | 72.099 | ENSUAMG00000011224 | BZW1 | 96 | 72.167 | Ursus_americanus |
| ENSG00000136261 | BZW2 | 96 | 94.037 | ENSUMAG00000022044 | BZW2 | 95 | 86.124 | Ursus_maritimus |
| ENSG00000136261 | BZW2 | 99 | 72.099 | ENSUMAG00000021441 | BZW1 | 77 | 72.167 | Ursus_maritimus |
| ENSG00000136261 | BZW2 | 99 | 71.875 | ENSVPAG00000006677 | BZW1 | 94 | 58.105 | Vicugna_pacos |
| ENSG00000136261 | BZW2 | 100 | 99.556 | ENSVPAG00000008439 | BZW2 | 100 | 95.704 | Vicugna_pacos |
| ENSG00000136261 | BZW2 | 100 | 100.000 | ENSVVUG00000008686 | BZW2 | 100 | 100.000 | Vulpes_vulpes |
| ENSG00000136261 | BZW2 | 99 | 72.099 | ENSVVUG00000019058 | BZW1 | 94 | 72.167 | Vulpes_vulpes |
| ENSG00000136261 | BZW2 | 99 | 72.050 | ENSXETG00000030733 | bzw1 | 96 | 70.443 | Xenopus_tropicalis |
| ENSG00000136261 | BZW2 | 100 | 89.951 | ENSXETG00000022795 | bzw2 | 97 | 89.951 | Xenopus_tropicalis |
| ENSG00000136261 | BZW2 | 99 | 71.429 | ENSXCOG00000016791 | bzw1a | 96 | 70.905 | Xiphophorus_couchianus |
| ENSG00000136261 | BZW2 | 99 | 68.944 | ENSXCOG00000014454 | - | 90 | 67.980 | Xiphophorus_couchianus |
| ENSG00000136261 | BZW2 | 99 | 68.944 | ENSXMAG00000010531 | - | 96 | 67.980 | Xiphophorus_maculatus |
| ENSG00000136261 | BZW2 | 99 | 71.429 | ENSXMAG00000026117 | bzw1a | 96 | 70.936 | Xiphophorus_maculatus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0005515 | protein binding | 25147208. | IPI | Function |
| GO:0005737 | cytoplasm | 21873635. | IBA | Component |
| GO:0005737 | cytoplasm | 25468996. | IDA | Component |
| GO:0007399 | nervous system development | - | IEA | Process |
| GO:0016020 | membrane | 19946888. | HDA | Component |
| GO:0030154 | cell differentiation | - | IEA | Process |
| GO:0045296 | cadherin binding | 25468996. | HDA | Function |
| PID | Title | Article | Time | Author | Doi |
|---|---|---|---|---|---|
| 28791373 |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| ACC | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| LAML | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PCPG | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| TGCT | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UVM | |||||||
| UVM |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| ACC | |||
| LUAD | |||
| PRAD | |||
| THCA |
| Cancer | P-value | Q-value |
|---|---|---|
| SARC | ||
| MESO | ||
| ACC | ||
| HNSC | ||
| SKCM | ||
| LUSC | ||
| KIRP | ||
| PAAD | ||
| CESC | ||
| KICH | ||
| UCEC | ||
| GBM | ||
| LIHC | ||
| LUAD | ||
| OV |