
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000412570 | SMPD4-203 | 3601 | - | ENSP00000403002 | 57 (aa) | - | F2Z2W5 |
| ENST00000482171 | SMPD4-218 | 4298 | - | - | - (aa) | - | - |
| ENST00000433118 | SMPD4-206 | 3335 | - | ENSP00000397278 | 56 (aa) | - | F2Z2I0 |
| ENST00000491319 | SMPD4-220 | 593 | - | - | - (aa) | - | - |
| ENST00000451542 | SMPD4-212 | 796 | - | ENSP00000405363 | 265 (aa) | - | H7C2E2 |
| ENST00000351288 | SMPD4-201 | 3661 | XM_011511444 | ENSP00000259217 | 837 (aa) | XP_011509746 | Q9NXE4 |
| ENST00000409031 | SMPD4-202 | 4896 | XM_011511443 | ENSP00000386531 | 866 (aa) | XP_011509745 | Q9NXE4 |
| ENST00000439886 | SMPD4-209 | 3371 | XM_011511445 | ENSP00000401648 | 741 (aa) | XP_011509747 | H7C1Q6 |
| ENST00000455548 | SMPD4-214 | 547 | - | - | - (aa) | - | - |
| ENST00000454468 | SMPD4-213 | 3586 | - | ENSP00000407591 | 50 (aa) | - | F8WF03 |
| ENST00000439029 | SMPD4-208 | 566 | - | ENSP00000416208 | 56 (aa) | - | F2Z2I0 |
| ENST00000473720 | SMPD4-217 | 555 | - | - | - (aa) | - | - |
| ENST00000431183 | SMPD4-205 | 3614 | - | ENSP00000405187 | 764 (aa) | - | Q9NXE4 |
| ENST00000457039 | SMPD4-215 | 940 | - | ENSP00000397078 | 313 (aa) | - | H7C0W5 |
| ENST00000461187 | SMPD4-216 | 659 | - | - | - (aa) | - | - |
| ENST00000491128 | SMPD4-219 | 2570 | - | - | - (aa) | - | - |
| ENST00000449159 | SMPD4-211 | 635 | - | ENSP00000403915 | 211 (aa) | - | H7C235 |
| ENST00000441135 | SMPD4-210 | 699 | - | ENSP00000406458 | 135 (aa) | - | C9J647 |
| ENST00000430682 | SMPD4-204 | 820 | - | ENSP00000406325 | 274 (aa) | - | H7C2J2 |
| ENST00000435455 | SMPD4-207 | 842 | - | ENSP00000393791 | 88 (aa) | - | F8WEQ5 |
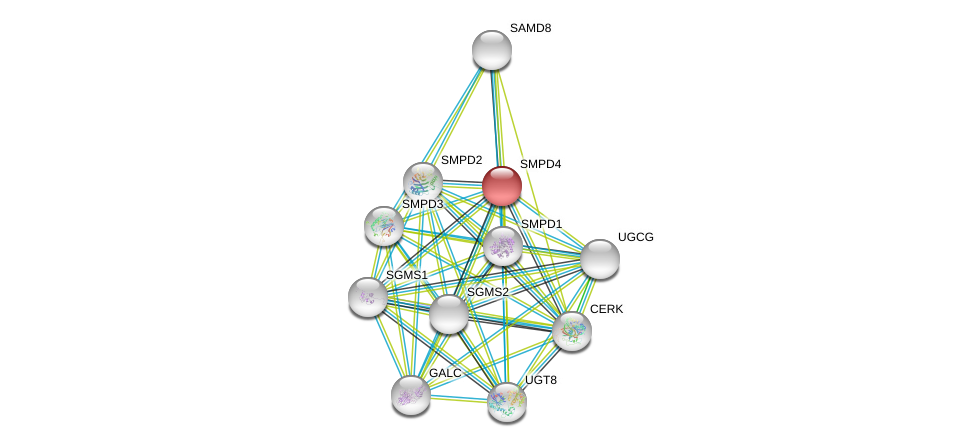
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000139 | Golgi membrane | - | IEA | Component |
| GO:0004767 | sphingomyelin phosphodiesterase activity | - | IEA | Function |
| GO:0005783 | endoplasmic reticulum | 16517606.18505924. | IDA | Component |
| GO:0005789 | endoplasmic reticulum membrane | - | TAS | Component |
| GO:0005794 | Golgi apparatus | 16517606. | IDA | Component |
| GO:0005802 | trans-Golgi network | 16517606. | IDA | Component |
| GO:0006685 | sphingomyelin catabolic process | 16517606. | IDA | Process |
| GO:0006687 | glycosphingolipid metabolic process | - | TAS | Process |
| GO:0016021 | integral component of membrane | - | IEA | Component |
| GO:0046475 | glycerophospholipid catabolic process | 16517606. | IDA | Process |
| GO:0046513 | ceramide biosynthetic process | 16517606. | IDA | Process |
| GO:0046872 | metal ion binding | - | IEA | Function |
| GO:0050290 | sphingomyelin phosphodiesterase D activity | 16517606. | IDA | Function |
| GO:0071356 | cellular response to tumor necrosis factor | 16517606. | IDA | Process |