
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000260665 | PPR | PF01535.20 | 3.1e-29 | 1 | 6 |
| ENSP00000260665 | PPR | PF01535.20 | 3.1e-29 | 2 | 6 |
| ENSP00000260665 | PPR | PF01535.20 | 3.1e-29 | 3 | 6 |
| ENSP00000260665 | PPR | PF01535.20 | 3.1e-29 | 4 | 6 |
| ENSP00000260665 | PPR | PF01535.20 | 3.1e-29 | 5 | 6 |
| ENSP00000260665 | PPR | PF01535.20 | 3.1e-29 | 6 | 6 |
| ENSP00000403637 | PPR | PF01535.20 | 1e-15 | 1 | 3 |
| ENSP00000403637 | PPR | PF01535.20 | 1e-15 | 2 | 3 |
| ENSP00000403637 | PPR | PF01535.20 | 1e-15 | 3 | 3 |
| ENSP00000386562 | PPR | PF01535.20 | 3.3e-15 | 1 | 3 |
| ENSP00000386562 | PPR | PF01535.20 | 3.3e-15 | 2 | 3 |
| ENSP00000386562 | PPR | PF01535.20 | 3.3e-15 | 3 | 3 |
| ENSP00000386234 | PPR | PF01535.20 | 3.6e-15 | 1 | 3 |
| ENSP00000386234 | PPR | PF01535.20 | 3.6e-15 | 2 | 3 |
| ENSP00000386234 | PPR | PF01535.20 | 3.6e-15 | 3 | 3 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000409659 | LRPPRC-202 | 1610 | - | ENSP00000386562 | 504 (aa) | - | B8ZZ38 |
| ENST00000419884 | LRPPRC-204 | 888 | - | ENSP00000414207 | 77 (aa) | - | H7C3W8 |
| ENST00000260665 | LRPPRC-201 | 6335 | XM_006711915 | ENSP00000260665 | 1394 (aa) | XP_006711978 | P42704 |
| ENST00000409946 | LRPPRC-203 | 1848 | - | ENSP00000386234 | 531 (aa) | - | A0A0C4DG06 |
| ENST00000483489 | LRPPRC-209 | 564 | - | - | - (aa) | - | - |
| ENST00000463456 | LRPPRC-206 | 3549 | - | - | - (aa) | - | - |
| ENST00000472420 | LRPPRC-208 | 694 | - | - | - (aa) | - | - |
| ENST00000447246 | LRPPRC-205 | 1209 | - | ENSP00000403637 | 359 (aa) | - | C9JCA9 |
| ENST00000467058 | LRPPRC-207 | 557 | - | - | - (aa) | - | - |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000138095 | Body Weight | 6.46942529539519E-5 | 17903300 |
| ENSG00000138095 | Body Weight | 4.40768165814054E-5 | 17903300 |
| ENSG00000138095 | Cholesterol, HDL | 7.84783516712504E-5 | 17903299 |
| ENSG00000138095 | Blood Glucose | 7.9570000E-005 | - |
| ENSG00000138095 | Platelet Function Tests | 1.0855100E-005 | - |
| ENSG00000138095 | Platelet Function Tests | 1.0845900E-005 | - |
| ENSG00000138095 | Child Development Disorders, Pervasive | 2.3762000E-005 | - |
| ENSG00000138095 | Intelligence | 9E-6 | 23358156 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000138095 | rs13387221 | 2 | 43908175 | A | Intelligence (childhood) | 23358156 | [0.035-0.093] unit increase | 0.064 | EFO_0004337 |
| ENSG00000138095 | rs4347883 | 2 | 43957990 | T | Cognitive performance | 30038396 | [0.01-0.022] unit decrease | 0.016 | EFO_0008354 |
| ENSG00000138095 | rs13017251 | 2 | 43994021 | A | Hand grip strength | 29691431 | [0.0014-0.0026] unit increase | 0.002 | EFO_0006941 |
| ENSG00000138095 | rs2955280 | 2 | 43889697 | T | Intelligence | 29942086 | z-score decrease | 5.455 | EFO_0004337 |
| ENSG00000138095 | rs1136998 | 2 | 43888044 | A | Smoking status (ever vs never smokers) | 30643258 | [0.014-0.026] unit decrease | 0.020011662 | EFO_0004318 |
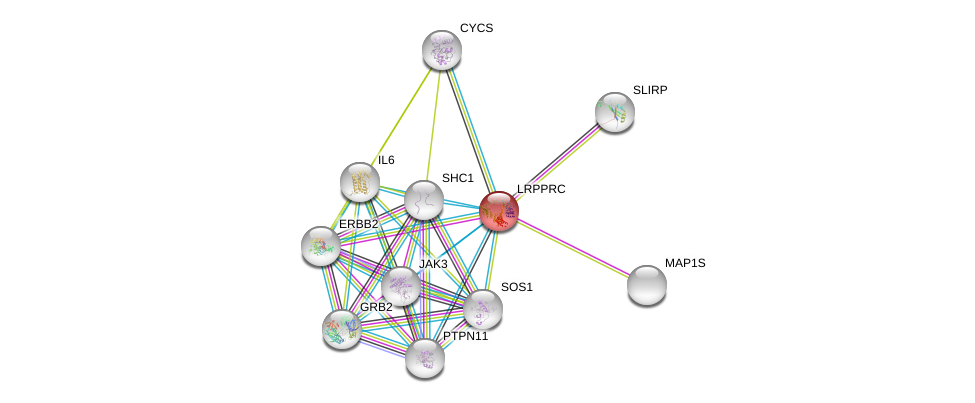
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000138095 | LRPPRC | 94 | 58.390 | ENSAPLG00000008902 | LRPPRC | 99 | 58.390 | Anas_platyrhynchos |
| ENSG00000138095 | LRPPRC | 99 | 80.871 | ENSBTAG00000016368 | LRPPRC | 99 | 80.532 | Bos_taurus |
| ENSG00000138095 | LRPPRC | 99 | 81.214 | ENSCAFG00000002527 | LRPPRC | 99 | 79.813 | Canis_familiaris |
| ENSG00000138095 | LRPPRC | 99 | 81.690 | ENSCAFG00020025995 | LRPPRC | 99 | 79.023 | Canis_lupus_dingo |
| ENSG00000138095 | LRPPRC | 93 | 87.164 | ENSTSYG00000013197 | LRPPRC | 96 | 83.631 | Carlito_syrichta |
| ENSG00000138095 | LRPPRC | 97 | 66.479 | ENSCPBG00000005446 | LRPPRC | 99 | 62.955 | Chrysemys_picta_bellii |
| ENSG00000138095 | LRPPRC | 87 | 45.714 | FBgn0284256 | bsf | 66 | 42.723 | Drosophila_melanogaster |
| ENSG00000138095 | LRPPRC | 97 | 85.632 | ENSEASG00005003715 | LRPPRC | 99 | 82.268 | Equus_asinus_asinus |
| ENSG00000138095 | LRPPRC | 99 | 83.106 | ENSECAG00000023814 | LRPPRC | 95 | 82.539 | Equus_caballus |
| ENSG00000138095 | LRPPRC | 91 | 65.152 | ENSGALG00000009967 | LRPPRC | 98 | 57.447 | Gallus_gallus |
| ENSG00000138095 | LRPPRC | 100 | 62.740 | ENSGAGG00000020227 | LRPPRC | 98 | 63.307 | Gopherus_agassizii |
| ENSG00000138095 | LRPPRC | 99 | 91.887 | ENSMLEG00000043673 | LRPPRC | 99 | 91.887 | Mandrillus_leucophaeus |
| ENSG00000138095 | LRPPRC | 96 | 57.797 | ENSMGAG00000010201 | LRPPRC | 100 | 57.797 | Meleagris_gallopavo |
| ENSG00000138095 | LRPPRC | 100 | 75.036 | MGP_CAROLIEiJ_G0021927 | Lrpprc | 100 | 75.036 | Mus_caroli |
| ENSG00000138095 | LRPPRC | 100 | 75.251 | ENSMUSG00000024120 | Lrpprc | 100 | 75.251 | Mus_musculus |
| ENSG00000138095 | LRPPRC | 100 | 76.255 | MGP_PahariEiJ_G0020917 | Lrpprc | 100 | 76.255 | Mus_pahari |
| ENSG00000138095 | LRPPRC | 100 | 75.251 | MGP_SPRETEiJ_G0022843 | Lrpprc | 100 | 75.251 | Mus_spretus |
| ENSG00000138095 | LRPPRC | 81 | 85.294 | ENSMEUG00000004430 | - | 88 | 85.294 | Notamacropus_eugenii |
| ENSG00000138095 | LRPPRC | 99 | 78.212 | ENSPEMG00000022720 | Lrpprc | 100 | 75.610 | Peromyscus_maniculatus_bairdii |
| ENSG00000138095 | LRPPRC | 95 | 74.499 | ENSPCIG00000006872 | LRPPRC | 99 | 67.906 | Phascolarctos_cinereus |
| ENSG00000138095 | LRPPRC | 99 | 84.034 | ENSPCOG00000001309 | LRPPRC | 99 | 83.992 | Propithecus_coquereli |
| ENSG00000138095 | LRPPRC | 97 | 93.333 | ENSRROG00000038362 | - | 87 | 88.793 | Rhinopithecus_roxellana |
| ENSG00000138095 | LRPPRC | 89 | 60.654 | ENSSPUG00000005188 | LRPPRC | 99 | 60.654 | Sphenodon_punctatus |
| ENSG00000138095 | LRPPRC | 78 | 75.000 | ENSTBEG00000011055 | - | 66 | 80.090 | Tupaia_belangeri |
| ENSG00000138095 | LRPPRC | 99 | 82.576 | ENSUAMG00000022940 | LRPPRC | 100 | 80.717 | Ursus_americanus |
| ENSG00000138095 | LRPPRC | 99 | 82.765 | ENSUMAG00000012801 | LRPPRC | 100 | 80.860 | Ursus_maritimus |
| ENSG00000138095 | LRPPRC | 99 | 81.214 | ENSVVUG00000002339 | LRPPRC | 99 | 78.879 | Vulpes_vulpes |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000794 | condensed nuclear chromosome | 12762840. | IDA | Component |
| GO:0000961 | negative regulation of mitochondrial RNA catabolic process | - | IEA | Process |
| GO:0003697 | single-stranded DNA binding | - | IEA | Function |
| GO:0003723 | RNA binding | 22658674.22681889. | HDA | Function |
| GO:0003723 | RNA binding | 12832482. | NAS | Function |
| GO:0005515 | protein binding | 15907802.17050673.19262567.22045337.26496610. | IPI | Function |
| GO:0005634 | nucleus | 12762840.12832482. | IDA | Component |
| GO:0005637 | nuclear inner membrane | - | IEA | Component |
| GO:0005640 | nuclear outer membrane | - | IEA | Component |
| GO:0005654 | nucleoplasm | - | IEA | Component |
| GO:0005739 | mitochondrion | 12832482. | IDA | Component |
| GO:0005739 | mitochondrion | 12762840. | IDA | Component |
| GO:0005856 | cytoskeleton | 12762840. | IDA | Component |
| GO:0005874 | microtubule | 21525035. | IDA | Component |
| GO:0008017 | microtubule binding | 12762840. | TAS | Function |
| GO:0016020 | membrane | 19946888. | HDA | Component |
| GO:0031625 | ubiquitin protein ligase binding | 19725078. | IPI | Function |
| GO:0042645 | mitochondrial nucleoid | 18063578. | IDA | Component |
| GO:0047497 | mitochondrion transport along microtubule | 12762840. | TAS | Process |
| GO:0048471 | perinuclear region of cytoplasm | 12762840. | IDA | Component |
| GO:0048487 | beta-tubulin binding | 12762840. | IDA | Function |
| GO:0051015 | actin filament binding | 12762840. | IDA | Function |
| GO:0051028 | mRNA transport | - | IEA | Process |
| GO:0070129 | regulation of mitochondrial translation | - | IEA | Process |
| GO:1990904 | ribonucleoprotein complex | - | IEA | Component |