
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000512542 | ANXA3-208 | 307 | - | ENSP00000426591 | 55 (aa) | - | D6RFJ9 |
| ENST00000512373 | ANXA3-207 | 545 | - | ENSP00000424584 | 104 (aa) | - | D6RAZ8 |
| ENST00000514171 | ANXA3-210 | 880 | - | ENSP00000421512 | 154 (aa) | - | D6RFG5 |
| ENST00000508214 | ANXA3-205 | 567 | - | ENSP00000422281 | 134 (aa) | - | D6RCA8 |
| ENST00000510502 | ANXA3-206 | 571 | - | - | - (aa) | - | - |
| ENST00000505805 | ANXA3-204 | 832 | - | - | - (aa) | - | - |
| ENST00000512884 | ANXA3-209 | 1335 | - | ENSP00000423068 | 284 (aa) | - | D6RA82 |
| ENST00000503776 | ANXA3-203 | 2932 | - | - | - (aa) | - | - |
| ENST00000503570 | ANXA3-202 | 1524 | - | ENSP00000421015 | 284 (aa) | - | D6RA82 |
| ENST00000264908 | ANXA3-201 | 1657 | - | ENSP00000264908 | 323 (aa) | - | P12429 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000138772 | rs7679218 | 4 | 78585710 | ? | Amyotrophic lateral sclerosis (sporadic) | 24529757 | EFO_0001357 | ||
| ENSG00000138772 | rs11335718 | 4 | 78607389 | A | Osteoarthritis (self-reported) | 29559693 | [1.07172-1.16949] | 1.11954 | EFO_0002506 |
| ENSG00000138772 | rs2867461 | 4 | 78592061 | A | Rheumatoid arthritis | 22446963 | [1.09-1.17] | 1.13 | EFO_0000685 |
| ENSG00000138772 | rs10028001 | 4 | 78581818 | A | Rheumatoid arthritis | 30423114 | [0.064-0.164] unit decrease | 0.11386 | EFO_0000685 |
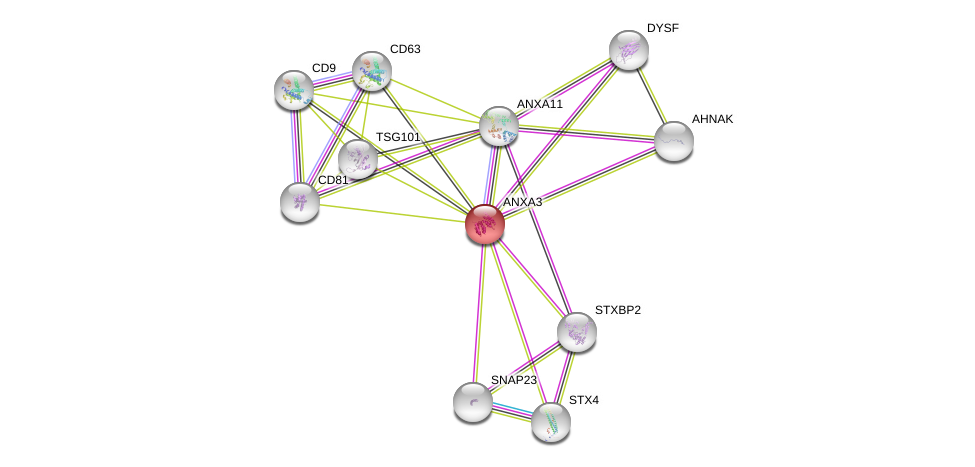
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0005509 | calcium ion binding | - | IEA | Function |
| GO:0005544 | calcium-dependent phospholipid binding | 21873635. | IBA | Function |
| GO:0005544 | calcium-dependent phospholipid binding | 2138016.2138632. | IDA | Function |
| GO:0005737 | cytoplasm | 17205602. | IDA | Component |
| GO:0005829 | cytosol | - | IDA | Component |
| GO:0005886 | plasma membrane | 21873635. | IBA | Component |
| GO:0005886 | plasma membrane | 7526843. | IDA | Component |
| GO:0006909 | phagocytosis | 21873635. | IBA | Process |
| GO:0006909 | phagocytosis | 9497492. | IDA | Process |
| GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules | 21873635. | IBA | Process |
| GO:0010595 | positive regulation of endothelial cell migration | 21873635. | IBA | Process |
| GO:0010595 | positive regulation of endothelial cell migration | 16236264. | IDA | Process |
| GO:0016020 | membrane | 17205602. | IDA | Component |
| GO:0019834 | phospholipase A2 inhibitor activity | 21873635. | IBA | Function |
| GO:0021766 | hippocampus development | - | IEA | Process |
| GO:0030424 | axon | - | IEA | Component |
| GO:0030425 | dendrite | - | IEA | Component |
| GO:0030670 | phagocytic vesicle membrane | 7526843. | IDA | Component |
| GO:0031100 | animal organ regeneration | - | IEA | Process |
| GO:0042581 | specific granule | 7526843. | IDA | Component |
| GO:0042742 | defense response to bacterium | 9497492. | IDA | Process |
| GO:0043025 | neuronal cell body | - | IEA | Component |
| GO:0043086 | negative regulation of catalytic activity | - | IEA | Process |
| GO:0043312 | neutrophil degranulation | 9550422. | IDA | Process |
| GO:0045766 | positive regulation of angiogenesis | 16236264. | IDA | Process |
| GO:0048306 | calcium-dependent protein binding | 21873635. | IBA | Function |
| GO:0048306 | calcium-dependent protein binding | 15226301. | IPI | Function |
| GO:0051054 | positive regulation of DNA metabolic process | - | IEA | Process |
| GO:0051091 | positive regulation of DNA-binding transcription factor activity | 16236264. | IDA | Process |
| GO:0051384 | response to glucocorticoid | 21873635. | IBA | Process |
| GO:0070062 | extracellular exosome | 19056867.23533145. | HDA | Component |
| GO:0070848 | response to growth factor | - | IEA | Process |
| GO:2000310 | regulation of NMDA receptor activity | 21873635. | IBA | Process |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| KICH | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| KIRP | |||||||
| KIRP | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| THCA | |||||||
| THCA | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| LIHC | |||
| LUAD | |||
| MESO |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| SARC | ||
| MESO | ||
| UCS | ||
| HNSC | ||
| ESCA | ||
| KIRP | ||
| COAD | ||
| PAAD | ||
| CESC | ||
| OV |