
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000512641 | PAPSS1-206 | 466 | - | - | - (aa) | - | - |
| ENST00000506544 | PAPSS1-204 | 577 | - | - | - (aa) | - | - |
| ENST00000265174 | PAPSS1-201 | 2731 | XM_011532400 | ENSP00000265174 | 624 (aa) | XP_011530702 | O43252 |
| ENST00000502431 | PAPSS1-202 | 903 | - | - | - (aa) | - | - |
| ENST00000514815 | PAPSS1-208 | 296 | - | - | - (aa) | - | - |
| ENST00000514489 | PAPSS1-207 | 579 | - | - | - (aa) | - | - |
| ENST00000511304 | PAPSS1-205 | 927 | - | - | - (aa) | - | - |
| ENST00000504987 | PAPSS1-203 | 492 | - | - | - (aa) | - | - |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000138801 | Stroke | 8.9481673E-004 | - |
| ENSG00000138801 | Erythrocyte Indices | 3.768e-005 | - |
| ENSG00000138801 | Arteries | 9.3540000E-005 | - |
| ENSG00000138801 | Coronary Disease | 5.1690000E-005 | - |
| ENSG00000138801 | Coronary Disease | 3.2850000E-005 | - |
| ENSG00000138801 | Coronary Disease | 2.3540000E-006 | - |
| ENSG00000138801 | Coronary Disease | 2.3540000E-006 | - |
| ENSG00000138801 | Coronary Disease | 3.0240000E-005 | - |
| ENSG00000138801 | Coronary Disease | 1.7980000E-005 | - |
| ENSG00000138801 | Coronary Disease | 2.0630000E-006 | - |
| ENSG00000138801 | Coronary Disease | 1.5070000E-006 | - |
| ENSG00000138801 | Coronary Disease | 8.6170000E-005 | - |
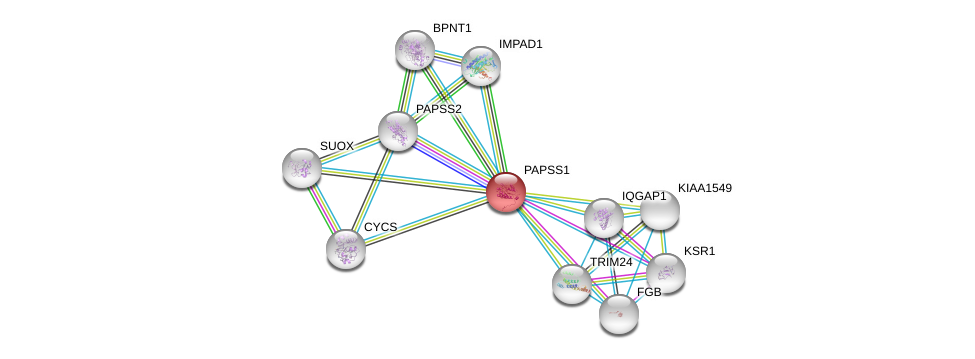
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000138801 | PAPSS1 | 96 | 78.369 | ENSG00000198682 | PAPSS2 | 98 | 78.369 | Homo_sapiens |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000103 | sulfate assimilation | 21873635. | IBA | Process |
| GO:0000103 | sulfate assimilation | 14747722. | IDA | Process |
| GO:0000103 | sulfate assimilation | - | IEA | Process |
| GO:0001501 | skeletal system development | 9771708. | TAS | Process |
| GO:0004020 | adenylylsulfate kinase activity | 21873635. | IBA | Function |
| GO:0004020 | adenylylsulfate kinase activity | 14747722.23207770. | IDA | Function |
| GO:0004781 | sulfate adenylyltransferase (ATP) activity | 14747722. | IDA | Function |
| GO:0004781 | sulfate adenylyltransferase (ATP) activity | - | ISS | Function |
| GO:0005524 | ATP binding | - | IEA | Function |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0016310 | phosphorylation | - | IEA | Process |
| GO:0016779 | nucleotidyltransferase activity | - | ISS | Function |
| GO:0042803 | protein homodimerization activity | 14747722. | IPI | Function |
| GO:0050428 | 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process | 21873635. | IBA | Process |
| GO:0050428 | 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process | 14747722.23207770. | IDA | Process |
| GO:0050428 | 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process | - | TAS | Process |