
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000267197 | RRM_1 | PF00076.22 | 8.2e-10 | 1 | 1 |
| ENSP00000442924 | RRM_1 | PF00076.22 | 8.2e-10 | 1 | 1 |
| ENSP00000474253 | RRM_1 | PF00076.22 | 8.4e-10 | 1 | 1 |
| ENSP00000481531 | RRM_1 | PF00076.22 | 8.4e-10 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000604567 | SETD1B-203 | 5969 | XM_005253858 | ENSP00000474253 | 1966 (aa) | XP_005253915 | Q9UPS6 |
| ENST00000542440 | SETD1B-202 | 8185 | - | ENSP00000442924 | 1923 (aa) | - | Q9UPS6 |
| ENST00000267197 | SETD1B-201 | 5772 | - | ENSP00000267197 | 1923 (aa) | - | A0A0A0MQV9 |
| ENST00000619791 | SETD1B-204 | 5901 | XM_006719296 | ENSP00000481531 | 1966 (aa) | XP_006719359 | Q9UPS6 |
| Pathway ID | Pathway Name | Source |
|---|---|---|
| hsa00310 | Lysine degradation | KEGG |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000139718 | Motor Activity | 3E-6 | 23251661 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000139718 | rs895953 | 12 | 121811142 | T | Coronary artery disease and HDL cholesterol levels (multivariate analysis) | 30525989 | EFO_0004612|EFO_0000378 | ||
| ENSG00000139718 | rs895953 | 12 | 121811142 | T | Coronary artery disease and triglyceride levels (multivariate analysis) | 30525989 | EFO_0004530|EFO_0000378 | ||
| ENSG00000139718 | rs7974348 | 12 | 121816807 | ? | White blood cell count | 30595370 | EFO_0004308 | ||
| ENSG00000139718 | rs895953 | 12 | 121811142 | ? | Lipid traits (pleiotropy) (HIPO component 1) | 30289880 | EFO_0004574|EFO_0004530|EFO_0004611|EFO_0004612 |
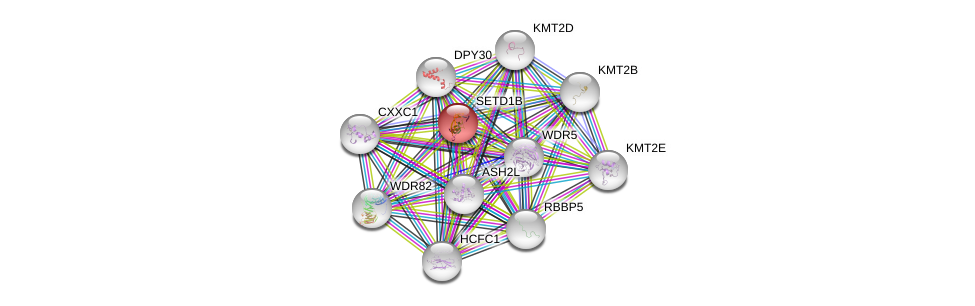
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000139718 | SETD1B | 66 | 82.595 | ENSAMEG00000014455 | SETD1B | 61 | 85.188 | Ailuropoda_melanoleuca |
| ENSG00000139718 | SETD1B | 71 | 61.300 | ENSACIG00000016453 | setd1ba | 54 | 48.187 | Amphilophus_citrinellus |
| ENSG00000139718 | SETD1B | 71 | 80.805 | ENSATEG00000021563 | setd1ba | 55 | 80.675 | Anabas_testudineus |
| ENSG00000139718 | SETD1B | 50 | 73.898 | ENSAPLG00000005217 | SETD1B | 70 | 75.000 | Anas_platyrhynchos |
| ENSG00000139718 | SETD1B | 76 | 97.037 | ENSANAG00000027480 | SETD1B | 61 | 91.702 | Aotus_nancymaae |
| ENSG00000139718 | SETD1B | 100 | 95.688 | ENSCJAG00000016930 | SETD1B | 100 | 95.688 | Callithrix_jacchus |
| ENSG00000139718 | SETD1B | 58 | 85.784 | ENSCAFG00020016619 | SETD1B | 57 | 97.153 | Canis_lupus_dingo |
| ENSG00000139718 | SETD1B | 72 | 86.769 | ENSCHIG00000013406 | SETD1B | 55 | 96.479 | Capra_hircus |
| ENSG00000139718 | SETD1B | 85 | 94.828 | ENSTSYG00000033777 | SETD1B | 68 | 97.727 | Carlito_syrichta |
| ENSG00000139718 | SETD1B | 100 | 96.156 | ENSCCAG00000031928 | SETD1B | 100 | 96.156 | Cebus_capucinus |
| ENSG00000139718 | SETD1B | 100 | 94.718 | ENSCATG00000035803 | SETD1B | 100 | 94.718 | Cercocebus_atys |
| ENSG00000139718 | SETD1B | 89 | 94.292 | ENSCSAG00000002187 | SETD1B | 98 | 93.265 | Chlorocebus_sabaeus |
| ENSG00000139718 | SETD1B | 88 | 96.496 | ENSCANG00000023445 | SETD1B | 100 | 96.496 | Colobus_angolensis_palliatus |
| ENSG00000139718 | SETD1B | 59 | 96.804 | ENSCGRG00000018075 | Setd1b | 80 | 96.379 | Cricetulus_griseus_crigri |
| ENSG00000139718 | SETD1B | 74 | 83.093 | ENSDNOG00000035084 | SETD1B | 98 | 81.568 | Dasypus_novemcinctus |
| ENSG00000139718 | SETD1B | 96 | 94.690 | ENSEASG00005012369 | SETD1B | 99 | 94.690 | Equus_asinus_asinus |
| ENSG00000139718 | SETD1B | 79 | 94.838 | ENSECAG00000006549 | SETD1B | 77 | 94.838 | Equus_caballus |
| ENSG00000139718 | SETD1B | 84 | 91.973 | ENSFDAG00000015120 | SETD1B | 62 | 92.540 | Fukomys_damarensis |
| ENSG00000139718 | SETD1B | 75 | 62.682 | ENSGALG00000004324 | SETD1B | 54 | 74.090 | Gallus_gallus |
| ENSG00000139718 | SETD1B | 86 | 56.162 | ENSGAFG00000008712 | setd1ba | 58 | 81.056 | Gambusia_affinis |
| ENSG00000139718 | SETD1B | 93 | 99.179 | ENSGGOG00000006715 | SETD1B | 100 | 92.223 | Gorilla_gorilla |
| ENSG00000139718 | SETD1B | 63 | 92.739 | ENSHGLG00000012063 | SETD1B | 70 | 91.883 | Heterocephalus_glaber_female |
| ENSG00000139718 | SETD1B | 83 | 57.302 | ENSKMAG00000007804 | setd1ba | 55 | 63.596 | Kryptolebias_marmoratus |
| ENSG00000139718 | SETD1B | 92 | 62.273 | ENSLACG00000018379 | SETD1B | 54 | 70.108 | Latimeria_chalumnae |
| ENSG00000139718 | SETD1B | 87 | 94.671 | ENSLAFG00000026518 | SETD1B | 95 | 94.671 | Loxodonta_africana |
| ENSG00000139718 | SETD1B | 100 | 93.601 | ENSMFAG00000030930 | SETD1B | 100 | 93.601 | Macaca_fascicularis |
| ENSG00000139718 | SETD1B | 100 | 94.806 | ENSMMUG00000012842 | SETD1B | 100 | 94.806 | Macaca_mulatta |
| ENSG00000139718 | SETD1B | 100 | 93.855 | ENSMNEG00000014228 | SETD1B | 100 | 93.855 | Macaca_nemestrina |
| ENSG00000139718 | SETD1B | 62 | 94.802 | ENSMLEG00000039573 | SETD1B | 63 | 95.215 | Mandrillus_leucophaeus |
| ENSG00000139718 | SETD1B | 63 | 62.285 | ENSMGAG00000004421 | SETD1B | 53 | 72.377 | Meleagris_gallopavo |
| ENSG00000139718 | SETD1B | 91 | 51.575 | ENSMMOG00000008190 | setd1ba | 50 | 81.505 | Mola_mola |
| ENSG00000139718 | SETD1B | 82 | 99.185 | ENSNLEG00000001254 | SETD1B | 53 | 99.178 | Nomascus_leucogenys |
| ENSG00000139718 | SETD1B | 76 | 93.064 | ENSODEG00000010155 | SETD1B | 82 | 93.931 | Octodon_degus |
| ENSG00000139718 | SETD1B | 95 | 86.381 | ENSOCUG00000017708 | SETD1B | 98 | 94.539 | Oryctolagus_cuniculus |
| ENSG00000139718 | SETD1B | 71 | 79.448 | ENSORLG00000008435 | setd1ba | 52 | 80.878 | Oryzias_latipes |
| ENSG00000139718 | SETD1B | 87 | 55.099 | ENSORLG00020017079 | setd1ba | 51 | 80.878 | Oryzias_latipes_hni |
| ENSG00000139718 | SETD1B | 67 | 79.077 | ENSOMEG00000002270 | setd1ba | 52 | 81.013 | Oryzias_melastigma |
| ENSG00000139718 | SETD1B | 62 | 84.811 | ENSOARG00000012777 | SETD1B | 63 | 84.799 | Ovis_aries |
| ENSG00000139718 | SETD1B | 100 | 96.748 | ENSPPAG00000043308 | SETD1B | 100 | 96.748 | Pan_paniscus |
| ENSG00000139718 | SETD1B | 89 | 95.568 | ENSPPRG00000021041 | SETD1B | 98 | 95.616 | Panthera_pardus |
| ENSG00000139718 | SETD1B | 100 | 99.339 | ENSPTRG00000005567 | SETD1B | 100 | 99.339 | Pan_troglodytes |
| ENSG00000139718 | SETD1B | 100 | 98.629 | ENSPANG00000014784 | SETD1B | 100 | 98.629 | Papio_anubis |
| ENSG00000139718 | SETD1B | 89 | 52.670 | ENSPMGG00000011527 | setd1ba | 50 | 72.554 | Periophthalmus_magnuspinnatus |
| ENSG00000139718 | SETD1B | 86 | 53.748 | ENSPFOG00000019768 | setd1ba | 50 | 82.019 | Poecilia_formosa |
| ENSG00000139718 | SETD1B | 91 | 52.304 | ENSPLAG00000023459 | setd1ba | 50 | 83.228 | Poecilia_latipinna |
| ENSG00000139718 | SETD1B | 90 | 53.352 | ENSPREG00000011589 | setd1ba | 50 | 83.228 | Poecilia_reticulata |
| ENSG00000139718 | SETD1B | 100 | 95.023 | ENSRBIG00000034516 | SETD1B | 100 | 95.023 | Rhinopithecus_bieti |
| ENSG00000139718 | SETD1B | 100 | 94.258 | ENSRROG00000011844 | SETD1B | 100 | 94.258 | Rhinopithecus_roxellana |
| ENSG00000139718 | SETD1B | 72 | 96.631 | ENSSBOG00000033322 | SETD1B | 100 | 96.631 | Saimiri_boliviensis_boliviensis |
| ENSG00000139718 | SETD1B | 70 | 95.265 | ENSSSCG00000009806 | SETD1B | 79 | 95.265 | Sus_scrofa |
| ENSG00000139718 | SETD1B | 57 | 91.349 | ENSUAMG00000008729 | SETD1B | 52 | 92.545 | Ursus_americanus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0003723 | RNA binding | - | IEA | Function |
| GO:0005515 | protein binding | 17998332. | IPI | Function |
| GO:0005654 | nucleoplasm | - | IDA | Component |
| GO:0005654 | nucleoplasm | - | TAS | Component |
| GO:0005694 | chromosome | - | IEA | Component |
| GO:0005829 | cytosol | - | IDA | Component |
| GO:0016607 | nuclear speck | - | IEA | Component |
| GO:0035097 | histone methyltransferase complex | 17355966. | IDA | Component |
| GO:0042800 | histone methyltransferase activity (H3-K4 specific) | 17355966. | IDA | Function |
| GO:0048188 | Set1C/COMPASS complex | 17998332.18838538. | IDA | Component |
| GO:0051568 | histone H3-K4 methylation | 17355966. | IDA | Process |