
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000354775 | ALDH9A1-201 | 2696 | XM_011509294 | ENSP00000346827 | 518 (aa) | XP_011507596 | P49189 |
| ENST00000461664 | ALDH9A1-202 | 899 | - | - | - (aa) | - | - |
| ENST00000463610 | ALDH9A1-203 | 559 | - | - | - (aa) | - | - |
| ENST00000491436 | ALDH9A1-205 | 819 | - | - | - (aa) | - | - |
| ENST00000471457 | ALDH9A1-204 | 568 | - | - | - (aa) | - | - |
| Pathway ID | Pathway Name | Source |
|---|---|---|
| hsa00010 | Glycolysis / Gluconeogenesis | KEGG |
| hsa00053 | Ascorbate and aldarate metabolism | KEGG |
| hsa00071 | Fatty acid degradation | KEGG |
| hsa00280 | Valine, leucine and isoleucine degradation | KEGG |
| hsa00310 | Lysine degradation | KEGG |
| hsa00330 | Arginine and proline metabolism | KEGG |
| hsa00340 | Histidine metabolism | KEGG |
| hsa00380 | Tryptophan metabolism | KEGG |
| hsa00410 | beta-Alanine metabolism | KEGG |
| hsa00561 | Glycerolipid metabolism | KEGG |
| hsa00620 | Pyruvate metabolism | KEGG |
| hsa01100 | Metabolic pathways | KEGG |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000143149 | Leukoaraiosis | 7.4391763E-005 | - |
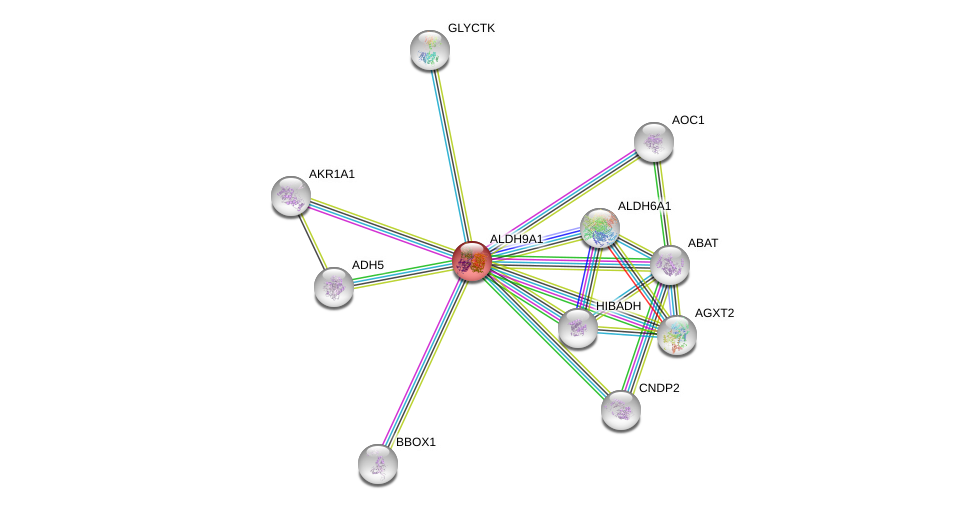
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0004029 | aldehyde dehydrogenase (NAD) activity | 21873635. | IBA | Function |
| GO:0004029 | aldehyde dehydrogenase (NAD) activity | 2925663. | IDA | Function |
| GO:0005737 | cytoplasm | 2925663. | TAS | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0006081 | cellular aldehyde metabolic process | 8645224. | IDA | Process |
| GO:0019145 | aminobutyraldehyde dehydrogenase activity | 21873635. | IBA | Function |
| GO:0019145 | aminobutyraldehyde dehydrogenase activity | 2925663.8645224. | IDA | Function |
| GO:0033737 | 1-pyrroline dehydrogenase activity | - | IEA | Function |
| GO:0042136 | neurotransmitter biosynthetic process | 2925663. | IDA | Process |
| GO:0042445 | hormone metabolic process | 2925663. | TAS | Process |
| GO:0043878 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (non-phosphorylating) activity | - | IEA | Function |
| GO:0045329 | carnitine biosynthetic process | - | IEA | Process |
| GO:0045329 | carnitine biosynthetic process | - | TAS | Process |
| GO:0047105 | 4-trimethylammoniobutyraldehyde dehydrogenase activity | 21873635. | IBA | Function |
| GO:0055114 | oxidation-reduction process | 2925663. | IDA | Process |
| GO:0070062 | extracellular exosome | 19056867.23533145. | HDA | Component |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| LGG | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| MESO | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SARC | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| DLBC | |||
| ESCA | |||
| KIRC | |||
| PAAD | |||
| SKCM | |||
| TGCT | |||
| THCA |
| Cancer | P-value | Q-value |
|---|---|---|
| THYM | ||
| KIRC | ||
| MESO | ||
| LUSC | ||
| PAAD | ||
| CESC | ||
| LGG | ||
| UVM |