
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000377523 | HA2 | PF04408.23 | 9.8e-18 | 1 | 1 |
| ENSP00000384621 | HA2 | PF04408.23 | 9.8e-18 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000404568 | DQX1-202 | 2689 | XM_011532645 | ENSP00000384621 | 717 (aa) | XP_011530947 | Q8TE96 |
| ENST00000495597 | DQX1-207 | 589 | - | - | - (aa) | - | - |
| ENST00000473508 | DQX1-205 | 2136 | - | - | - (aa) | - | - |
| ENST00000498552 | DQX1-208 | 100 | - | - | - (aa) | - | - |
| ENST00000483555 | DQX1-206 | 832 | - | - | - (aa) | - | - |
| ENST00000451518 | DQX1-204 | 555 | - | ENSP00000392969 | 119 (aa) | - | C9J0W1 |
| ENST00000393951 | DQX1-201 | 2540 | - | ENSP00000377523 | 717 (aa) | - | Q8TE96 |
| ENST00000418139 | DQX1-203 | 1688 | - | ENSP00000389196 | 132 (aa) | - | H7BZE3 |
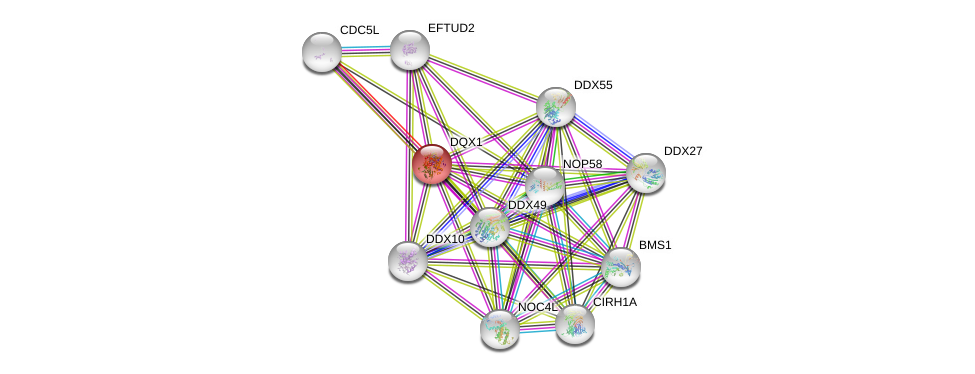
| Ensembl ID | Gene Symbol | Coverage | Identiy | Paralog | Gene Symbol | Coverage | Identiy |
|---|---|---|---|---|---|---|---|
| ENSG00000144045 | DQX1 | 100 | 35.833 | ENSG00000204560 | DHX16 | 90 | 31.538 |
| ENSG00000144045 | DQX1 | 93 | 32.362 | ENSG00000101452 | DHX35 | 93 | 32.362 |
| ENSG00000144045 | DQX1 | 93 | 38.248 | ENSG00000109606 | DHX15 | 84 | 38.248 |
| ENSG00000144045 | DQX1 | 91 | 30.643 | ENSG00000005100 | DHX33 | 91 | 30.344 |
| ENSG00000144045 | DQX1 | 90 | 40.187 | ENSG00000140829 | DHX38 | 71 | 34.409 |
| ENSG00000144045 | DQX1 | 95 | 44.209 | ENSG00000089876 | DHX32 | 95 | 49.148 |
| ENSG00000144045 | DQX1 | 90 | 38.182 | ENSG00000108406 | DHX40 | 87 | 35.391 |
| ENSG00000144045 | DQX1 | 73 | 30.418 | ENSG00000150990 | DHX37 | 58 | 30.418 |
| ENSG00000144045 | DQX1 | 91 | 32.675 | ENSG00000067596 | DHX8 | 52 | 32.675 |
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000144045 | DQX1 | 97 | 51.944 | ENSAPOG00000005067 | dqx1 | 91 | 51.719 | Acanthochromis_polyacanthus |
| ENSG00000144045 | DQX1 | 100 | 88.317 | ENSAMEG00000017193 | DQX1 | 100 | 88.317 | Ailuropoda_melanoleuca |
| ENSG00000144045 | DQX1 | 51 | 55.645 | ENSACIG00000007128 | - | 95 | 55.645 | Amphilophus_citrinellus |
| ENSG00000144045 | DQX1 | 97 | 51.857 | ENSAOCG00000020792 | dqx1 | 91 | 51.635 | Amphiprion_ocellaris |
| ENSG00000144045 | DQX1 | 97 | 51.872 | ENSAPEG00000011785 | dqx1 | 91 | 51.648 | Amphiprion_percula |
| ENSG00000144045 | DQX1 | 98 | 48.708 | ENSATEG00000009375 | dqx1 | 94 | 48.708 | Anabas_testudineus |
| ENSG00000144045 | DQX1 | 86 | 60.160 | ENSACAG00000004745 | DQX1 | 94 | 60.160 | Anolis_carolinensis |
| ENSG00000144045 | DQX1 | 100 | 94.730 | ENSANAG00000033980 | DQX1 | 100 | 94.730 | Aotus_nancymaae |
| ENSG00000144045 | DQX1 | 97 | 51.377 | ENSACLG00000006845 | dqx1 | 91 | 51.377 | Astatotilapia_calliptera |
| ENSG00000144045 | DQX1 | 95 | 53.073 | ENSAMXG00000009153 | dqx1 | 91 | 53.073 | Astyanax_mexicanus |
| ENSG00000144045 | DQX1 | 100 | 86.092 | ENSBTAG00000016609 | DQX1 | 100 | 86.092 | Bos_taurus |
| ENSG00000144045 | DQX1 | 100 | 94.576 | ENSCJAG00000017388 | DQX1 | 100 | 94.576 | Callithrix_jacchus |
| ENSG00000144045 | DQX1 | 100 | 87.761 | ENSCAFG00000008425 | DQX1 | 100 | 87.761 | Canis_familiaris |
| ENSG00000144045 | DQX1 | 99 | 85.593 | ENSCAFG00020014095 | DQX1 | 99 | 85.160 | Canis_lupus_dingo |
| ENSG00000144045 | DQX1 | 100 | 86.787 | ENSCHIG00000019357 | DQX1 | 100 | 86.787 | Capra_hircus |
| ENSG00000144045 | DQX1 | 100 | 84.284 | ENSTSYG00000001608 | DQX1 | 100 | 84.284 | Carlito_syrichta |
| ENSG00000144045 | DQX1 | 93 | 77.928 | ENSCAPG00000000387 | DQX1 | 97 | 77.928 | Cavia_aperea |
| ENSG00000144045 | DQX1 | 100 | 87.204 | ENSCPOG00000022849 | DQX1 | 100 | 87.204 | Cavia_porcellus |
| ENSG00000144045 | DQX1 | 100 | 95.828 | ENSCCAG00000031484 | DQX1 | 100 | 95.828 | Cebus_capucinus |
| ENSG00000144045 | DQX1 | 100 | 97.350 | ENSCATG00000037156 | DQX1 | 100 | 97.350 | Cercocebus_atys |
| ENSG00000144045 | DQX1 | 100 | 88.039 | ENSCLAG00000007521 | DQX1 | 100 | 88.039 | Chinchilla_lanigera |
| ENSG00000144045 | DQX1 | 100 | 97.211 | ENSCSAG00000015682 | DQX1 | 100 | 97.211 | Chlorocebus_sabaeus |
| ENSG00000144045 | DQX1 | 94 | 63.188 | ENSCPBG00000024467 | DQX1 | 86 | 63.188 | Chrysemys_picta_bellii |
| ENSG00000144045 | DQX1 | 100 | 96.932 | ENSCANG00000039725 | DQX1 | 100 | 96.932 | Colobus_angolensis_palliatus |
| ENSG00000144045 | DQX1 | 99 | 85.112 | ENSCGRG00001011093 | Dqx1 | 99 | 85.112 | Cricetulus_griseus_chok1gshd |
| ENSG00000144045 | DQX1 | 99 | 85.112 | ENSCGRG00000015486 | Dqx1 | 99 | 85.112 | Cricetulus_griseus_crigri |
| ENSG00000144045 | DQX1 | 80 | 48.336 | ENSCSEG00000018927 | dqx1 | 96 | 48.336 | Cynoglossus_semilaevis |
| ENSG00000144045 | DQX1 | 97 | 50.067 | ENSCVAG00000007365 | dqx1 | 94 | 49.934 | Cyprinodon_variegatus |
| ENSG00000144045 | DQX1 | 100 | 84.979 | ENSDNOG00000019907 | DQX1 | 100 | 84.979 | Dasypus_novemcinctus |
| ENSG00000144045 | DQX1 | 100 | 87.917 | ENSDORG00000013672 | Dqx1 | 100 | 87.917 | Dipodomys_ordii |
| ENSG00000144045 | DQX1 | 99 | 81.356 | ENSETEG00000011890 | DQX1 | 100 | 84.385 | Echinops_telfairi |
| ENSG00000144045 | DQX1 | 98 | 45.526 | ENSEBUG00000005072 | DQX1 | 72 | 45.526 | Eptatretus_burgeri |
| ENSG00000144045 | DQX1 | 99 | 88.999 | ENSEASG00005010654 | DQX1 | 99 | 88.999 | Equus_asinus_asinus |
| ENSG00000144045 | DQX1 | 99 | 88.999 | ENSECAG00000023896 | DQX1 | 99 | 88.999 | Equus_caballus |
| ENSG00000144045 | DQX1 | 96 | 51.018 | ENSELUG00000015619 | dqx1 | 91 | 51.018 | Esox_lucius |
| ENSG00000144045 | DQX1 | 100 | 89.152 | ENSFCAG00000028272 | DQX1 | 100 | 89.152 | Felis_catus |
| ENSG00000144045 | DQX1 | 94 | 61.674 | ENSFALG00000010921 | DQX1 | 96 | 59.888 | Ficedula_albicollis |
| ENSG00000144045 | DQX1 | 100 | 88.595 | ENSFDAG00000012179 | DQX1 | 100 | 88.595 | Fukomys_damarensis |
| ENSG00000144045 | DQX1 | 97 | 49.930 | ENSGMOG00000008923 | dqx1 | 99 | 49.728 | Gadus_morhua |
| ENSG00000144045 | DQX1 | 98 | 60.306 | ENSGALG00000033656 | DQX1 | 97 | 59.972 | Gallus_gallus |
| ENSG00000144045 | DQX1 | 97 | 49.134 | ENSGAFG00000008279 | dqx1 | 96 | 48.695 | Gambusia_affinis |
| ENSG00000144045 | DQX1 | 97 | 50.413 | ENSGACG00000018495 | dqx1 | 98 | 50.136 | Gasterosteus_aculeatus |
| ENSG00000144045 | DQX1 | 100 | 100.000 | ENSGGOG00000006307 | DQX1 | 100 | 99.442 | Gorilla_gorilla |
| ENSG00000144045 | DQX1 | 97 | 51.377 | ENSHBUG00000012890 | dqx1 | 91 | 51.377 | Haplochromis_burtoni |
| ENSG00000144045 | DQX1 | 99 | 87.288 | ENSHGLG00000011994 | DQX1 | 99 | 80.559 | Heterocephalus_glaber_female |
| ENSG00000144045 | DQX1 | 99 | 87.288 | ENSHGLG00100012005 | DQX1 | 99 | 80.559 | Heterocephalus_glaber_male |
| ENSG00000144045 | DQX1 | 97 | 50.392 | ENSHCOG00000002562 | dqx1 | 94 | 50.392 | Hippocampus_comes |
| ENSG00000144045 | DQX1 | 97 | 52.797 | ENSIPUG00000015152 | DQX1 | 95 | 52.797 | Ictalurus_punctatus |
| ENSG00000144045 | DQX1 | 100 | 88.178 | ENSSTOG00000026252 | DQX1 | 100 | 88.178 | Ictidomys_tridecemlineatus |
| ENSG00000144045 | DQX1 | 99 | 86.779 | ENSJJAG00000017070 | Dqx1 | 99 | 86.779 | Jaculus_jaculus |
| ENSG00000144045 | DQX1 | 94 | 52.941 | ENSKMAG00000002776 | dqx1 | 89 | 53.058 | Kryptolebias_marmoratus |
| ENSG00000144045 | DQX1 | 97 | 50.954 | ENSLBEG00000020503 | dqx1 | 95 | 50.670 | Labrus_bergylta |
| ENSG00000144045 | DQX1 | 95 | 46.262 | ENSLACG00000007417 | - | 95 | 45.135 | Latimeria_chalumnae |
| ENSG00000144045 | DQX1 | 90 | 48.148 | ENSLOCG00000001140 | dqx1 | 100 | 55.054 | Lepisosteus_oculatus |
| ENSG00000144045 | DQX1 | 99 | 88.128 | ENSLAFG00000018370 | DQX1 | 100 | 88.128 | Loxodonta_africana |
| ENSG00000144045 | DQX1 | 100 | 97.211 | ENSMFAG00000001892 | DQX1 | 100 | 97.211 | Macaca_fascicularis |
| ENSG00000144045 | DQX1 | 100 | 97.071 | ENSMMUG00000002129 | DQX1 | 100 | 97.071 | Macaca_mulatta |
| ENSG00000144045 | DQX1 | 100 | 97.350 | ENSMNEG00000043354 | DQX1 | 100 | 97.350 | Macaca_nemestrina |
| ENSG00000144045 | DQX1 | 100 | 97.350 | ENSMLEG00000028743 | DQX1 | 100 | 97.350 | Mandrillus_leucophaeus |
| ENSG00000144045 | DQX1 | 94 | 51.440 | ENSMAMG00000022633 | dqx1 | 94 | 50.344 | Mastacembelus_armatus |
| ENSG00000144045 | DQX1 | 97 | 51.377 | ENSMZEG00005002422 | dqx1 | 91 | 51.377 | Maylandia_zebra |
| ENSG00000144045 | DQX1 | 98 | 84.495 | ENSMAUG00000022113 | Dqx1 | 99 | 84.495 | Mesocricetus_auratus |
| ENSG00000144045 | DQX1 | 100 | 91.516 | ENSMICG00000006232 | DQX1 | 100 | 91.516 | Microcebus_murinus |
| ENSG00000144045 | DQX1 | 99 | 83.613 | ENSMOCG00000017871 | Dqx1 | 99 | 83.613 | Microtus_ochrogaster |
| ENSG00000144045 | DQX1 | 97 | 50.536 | ENSMMOG00000003933 | dqx1 | 94 | 50.265 | Mola_mola |
| ENSG00000144045 | DQX1 | 97 | 76.338 | ENSMODG00000004879 | DQX1 | 100 | 77.966 | Monodelphis_domestica |
| ENSG00000144045 | DQX1 | 94 | 50.822 | ENSMALG00000015132 | dqx1 | 90 | 50.403 | Monopterus_albus |
| ENSG00000144045 | DQX1 | 99 | 84.454 | ENSMUSG00000009145 | Dqx1 | 99 | 84.454 | Mus_musculus |
| ENSG00000144045 | DQX1 | 99 | 86.441 | ENSNGAG00000022609 | Dqx1 | 99 | 86.115 | Nannospalax_galili |
| ENSG00000144045 | DQX1 | 97 | 51.377 | ENSNBRG00000004923 | dqx1 | 91 | 51.377 | Neolamprologus_brichardi |
| ENSG00000144045 | DQX1 | 100 | 96.513 | ENSNLEG00000000771 | DQX1 | 100 | 96.513 | Nomascus_leucogenys |
| ENSG00000144045 | DQX1 | 99 | 72.034 | ENSMEUG00000013231 | DQX1 | 100 | 72.082 | Notamacropus_eugenii |
| ENSG00000144045 | DQX1 | 100 | 87.065 | ENSODEG00000012521 | DQX1 | 100 | 87.065 | Octodon_degus |
| ENSG00000144045 | DQX1 | 97 | 51.950 | ENSONIG00000012572 | dqx1 | 96 | 51.950 | Oreochromis_niloticus |
| ENSG00000144045 | DQX1 | 99 | 87.288 | ENSOCUG00000015372 | DQX1 | 100 | 82.749 | Oryctolagus_cuniculus |
| ENSG00000144045 | DQX1 | 94 | 51.909 | ENSORLG00000014277 | dqx1 | 92 | 51.163 | Oryzias_latipes |
| ENSG00000144045 | DQX1 | 94 | 51.841 | ENSORLG00020004148 | dqx1 | 92 | 51.096 | Oryzias_latipes_hni |
| ENSG00000144045 | DQX1 | 94 | 52.192 | ENSORLG00015017580 | dqx1 | 89 | 52.113 | Oryzias_latipes_hsok |
| ENSG00000144045 | DQX1 | 97 | 50.538 | ENSOMEG00000015481 | dqx1 | 95 | 50.469 | Oryzias_melastigma |
| ENSG00000144045 | DQX1 | 100 | 89.708 | ENSOGAG00000001065 | DQX1 | 100 | 89.708 | Otolemur_garnettii |
| ENSG00000144045 | DQX1 | 100 | 87.204 | ENSOARG00000012775 | DQX1 | 99 | 87.204 | Ovis_aries |
| ENSG00000144045 | DQX1 | 100 | 100.000 | ENSPPAG00000034827 | DQX1 | 100 | 96.585 | Pan_paniscus |
| ENSG00000144045 | DQX1 | 100 | 88.873 | ENSPPRG00000007947 | DQX1 | 100 | 88.873 | Panthera_pardus |
| ENSG00000144045 | DQX1 | 100 | 88.178 | ENSPTIG00000016302 | DQX1 | 100 | 88.178 | Panthera_tigris_altaica |
| ENSG00000144045 | DQX1 | 100 | 100.000 | ENSPTRG00000012096 | DQX1 | 100 | 99.582 | Pan_troglodytes |
| ENSG00000144045 | DQX1 | 100 | 97.211 | ENSPANG00000014436 | DQX1 | 100 | 97.211 | Papio_anubis |
| ENSG00000144045 | DQX1 | 96 | 56.835 | ENSPKIG00000016273 | dqx1 | 93 | 49.584 | Paramormyrops_kingsleyae |
| ENSG00000144045 | DQX1 | 100 | 85.160 | ENSPEMG00000000695 | Dqx1 | 100 | 85.160 | Peromyscus_maniculatus_bairdii |
| ENSG00000144045 | DQX1 | 89 | 46.226 | ENSPMAG00000000247 | dqx1 | 99 | 50.292 | Petromyzon_marinus |
| ENSG00000144045 | DQX1 | 99 | 77.731 | ENSPCIG00000006057 | DQX1 | 99 | 77.731 | Phascolarctos_cinereus |
| ENSG00000144045 | DQX1 | 97 | 50.203 | ENSPFOG00000006654 | dqx1 | 97 | 49.735 | Poecilia_formosa |
| ENSG00000144045 | DQX1 | 97 | 50.203 | ENSPLAG00000019886 | dqx1 | 97 | 49.735 | Poecilia_latipinna |
| ENSG00000144045 | DQX1 | 96 | 50.754 | ENSPMEG00000004991 | dqx1 | 88 | 50.269 | Poecilia_mexicana |
| ENSG00000144045 | DQX1 | 94 | 50.989 | ENSPREG00000007179 | dqx1 | 89 | 49.734 | Poecilia_reticulata |
| ENSG00000144045 | DQX1 | 100 | 100.000 | ENSPPYG00000012268 | DQX1 | 100 | 98.664 | Pongo_abelii |
| ENSG00000144045 | DQX1 | 84 | 85.839 | ENSPCAG00000016943 | DQX1 | 84 | 85.839 | Procavia_capensis |
| ENSG00000144045 | DQX1 | 100 | 91.377 | ENSPCOG00000021586 | DQX1 | 100 | 91.377 | Propithecus_coquereli |
| ENSG00000144045 | DQX1 | 100 | 87.344 | ENSPVAG00000013964 | DQX1 | 100 | 87.344 | Pteropus_vampyrus |
| ENSG00000144045 | DQX1 | 97 | 51.240 | ENSPNYG00000003028 | dqx1 | 91 | 51.240 | Pundamilia_nyererei |
| ENSG00000144045 | DQX1 | 96 | 53.138 | ENSPNAG00000023120 | dqx1 | 92 | 53.138 | Pygocentrus_nattereri |
| ENSG00000144045 | DQX1 | 99 | 84.712 | ENSRNOG00000008062 | Dqx1 | 99 | 84.712 | Rattus_norvegicus |
| ENSG00000144045 | DQX1 | 100 | 97.071 | ENSRBIG00000041823 | DQX1 | 100 | 97.071 | Rhinopithecus_bieti |
| ENSG00000144045 | DQX1 | 100 | 97.071 | ENSRROG00000039342 | DQX1 | 100 | 97.071 | Rhinopithecus_roxellana |
| ENSG00000144045 | DQX1 | 100 | 95.132 | ENSSBOG00000028107 | DQX1 | 100 | 95.132 | Saimiri_boliviensis_boliviensis |
| ENSG00000144045 | DQX1 | 99 | 77.560 | ENSSHAG00000016833 | DQX1 | 99 | 77.560 | Sarcophilus_harrisii |
| ENSG00000144045 | DQX1 | 95 | 50.354 | ENSSFOG00015003817 | dqx1 | 91 | 50.282 | Scleropages_formosus |
| ENSG00000144045 | DQX1 | 97 | 48.100 | ENSSMAG00000010612 | dqx1 | 94 | 48.114 | Scophthalmus_maximus |
| ENSG00000144045 | DQX1 | 94 | 50.548 | ENSSDUG00000020759 | dqx1 | 91 | 50.548 | Seriola_dumerili |
| ENSG00000144045 | DQX1 | 94 | 50.685 | ENSSLDG00000022712 | dqx1 | 91 | 50.685 | Seriola_lalandi_dorsalis |
| ENSG00000144045 | DQX1 | 98 | 51.715 | ENSSPAG00000013343 | dqx1 | 94 | 51.715 | Stegastes_partitus |
| ENSG00000144045 | DQX1 | 100 | 89.708 | ENSSSCG00000022614 | DQX1 | 100 | 89.708 | Sus_scrofa |
| ENSG00000144045 | DQX1 | 100 | 46.509 | ENSTRUG00000012911 | dqx1 | 100 | 51.554 | Takifugu_rubripes |
| ENSG00000144045 | DQX1 | 99 | 84.746 | ENSTBEG00000010252 | DQX1 | 100 | 87.978 | Tupaia_belangeri |
| ENSG00000144045 | DQX1 | 82 | 89.933 | ENSTTRG00000012725 | DQX1 | 82 | 89.933 | Tursiops_truncatus |
| ENSG00000144045 | DQX1 | 100 | 87.761 | ENSUAMG00000021690 | DQX1 | 100 | 87.761 | Ursus_americanus |
| ENSG00000144045 | DQX1 | 100 | 86.926 | ENSUMAG00000001677 | DQX1 | 100 | 86.926 | Ursus_maritimus |
| ENSG00000144045 | DQX1 | 50 | 92.199 | ENSVPAG00000007907 | - | 50 | 92.199 | Vicugna_pacos |
| ENSG00000144045 | DQX1 | 100 | 88.039 | ENSVVUG00000004178 | DQX1 | 100 | 88.039 | Vulpes_vulpes |
| ENSG00000144045 | DQX1 | 97 | 49.933 | ENSXMAG00000010692 | dqx1 | 90 | 49.472 | Xiphophorus_maculatus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0003723 | RNA binding | 21873635. | IBA | Function |
| GO:0005524 | ATP binding | - | IEA | Function |
| GO:0005634 | nucleus | - | IEA | Component |
| GO:0034459 | ATP-dependent 3'-5' RNA helicase activity | 21873635. | IBA | Function |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| ACC | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| LAML | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SARC | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| KIRP | |||
| PAAD | |||
| READ |
| Cancer | P-value | Q-value |
|---|---|---|
| MESO | ||
| ACC | ||
| SKCM | ||
| PRAD | ||
| LUSC | ||
| KIRP | ||
| BLCA | ||
| LIHC | ||
| DLBC | ||
| UVM |