
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000474665 | LIMD1-204 | 513 | - | - | - (aa) | - | - |
| ENST00000465039 | LIMD1-203 | 676 | - | - | - (aa) | - | - |
| ENST00000273317 | LIMD1-201 | 11331 | - | ENSP00000273317 | 676 (aa) | - | Q9UGP4 |
| ENST00000440097 | LIMD1-202 | 2773 | - | ENSP00000394537 | 620 (aa) | - | C9JRJ5 |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000144791 | Platelet Function Tests | 3.0728000E-006 | - |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000144791 | rs116425179 | 3 | 45557211 | G | Monocyte chemoattractant protein-1 levels | 27989323 | [0.098-0.198] SD units increase | 0.1476 | EFO_0004749 |
| ENSG00000144791 | rs34877991 | 3 | 45602156 | C | Systolic blood pressure | 30578418 | [0.12-0.23] mmHg decrease | 0.1731 | EFO_0006335 |
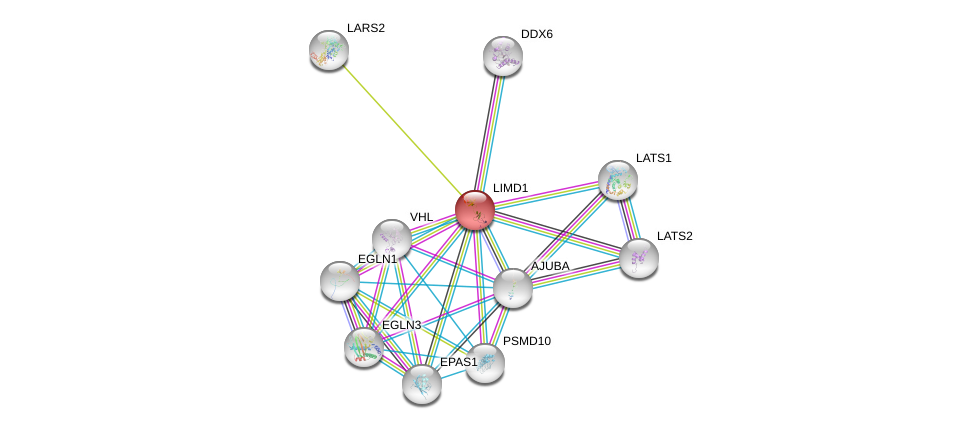
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000932 | P-body | 21873635. | IBA | Component |
| GO:0000932 | P-body | 20616046. | IDA | Component |
| GO:0001666 | response to hypoxia | 21873635. | IBA | Process |
| GO:0001666 | response to hypoxia | 22286099. | IDA | Process |
| GO:0002076 | osteoblast development | - | ISS | Process |
| GO:0003714 | transcription corepressor activity | 21873635. | IBA | Function |
| GO:0003714 | transcription corepressor activity | - | ISS | Function |
| GO:0005515 | protein binding | 15542589.18331720.20303269.20616046.22190034.22286099. | IPI | Function |
| GO:0005634 | nucleus | 21873635. | IBA | Component |
| GO:0005634 | nucleus | 15542589.18439753. | IDA | Component |
| GO:0005667 | transcription factor complex | 21873635. | IBA | Component |
| GO:0005737 | cytoplasm | 15542589.18439753. | IDA | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0005886 | plasma membrane | - | IDA | Component |
| GO:0005911 | cell-cell junction | 21873635. | IBA | Component |
| GO:0005912 | adherens junction | 21873635. | IBA | Component |
| GO:0005912 | adherens junction | 20303269. | IDA | Component |
| GO:0005925 | focal adhesion | 18439753. | IDA | Component |
| GO:0006355 | regulation of transcription, DNA-templated | 10647888. | TAS | Process |
| GO:0007010 | cytoskeleton organization | 21873635. | IBA | Process |
| GO:0007010 | cytoskeleton organization | 21834987. | IMP | Process |
| GO:0007165 | signal transduction | 10647888. | TAS | Process |
| GO:0007275 | multicellular organism development | 10647888. | TAS | Process |
| GO:0008270 | zinc ion binding | 10647888. | TAS | Function |
| GO:0008360 | regulation of cell shape | 21834987. | IMP | Process |
| GO:0016310 | phosphorylation | 18439753. | IDA | Process |
| GO:0016442 | RISC complex | 20616046. | IDA | Component |
| GO:0016477 | cell migration | 21834987. | IMP | Process |
| GO:0033962 | cytoplasmic mRNA processing body assembly | 20616046. | IMP | Process |
| GO:0035195 | gene silencing by miRNA | 20616046. | IMP | Process |
| GO:0035331 | negative regulation of hippo signaling | 21873635. | IBA | Process |
| GO:0035331 | negative regulation of hippo signaling | 20303269. | IDA | Process |
| GO:0045668 | negative regulation of osteoblast differentiation | - | ISS | Process |
| GO:0045892 | negative regulation of transcription, DNA-templated | 15542589. | IDA | Process |
| GO:0045892 | negative regulation of transcription, DNA-templated | 22286099. | IMP | Process |
| GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia | - | TAS | Process |
| GO:0090090 | negative regulation of canonical Wnt signaling pathway | - | ISS | Process |
| GO:2000637 | positive regulation of gene silencing by miRNA | 21873635. | IBA | Process |
| GO:2000637 | positive regulation of gene silencing by miRNA | 20616046. | IMP | Process |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CHOL | |||||||
| CHOL | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| DLBC | |||||||
| DLBC | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| KIRP | |||||||
| LAML | |||||||
| LAML | |||||||
| LGG | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| READ | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| THCA | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| CHOL | |||
| DLBC | |||
| GBM | |||
| LGG | |||
| LUAD | |||
| PAAD | |||
| SARC | |||
| SKCM | |||
| TGCT | |||
| THYM |
| Cancer | P-value | Q-value |
|---|---|---|
| THYM | ||
| KIRC | ||
| STAD | ||
| SARC | ||
| HNSC | ||
| PRAD | ||
| KIRP | ||
| BLCA | ||
| READ | ||
| KICH | ||
| LGG | ||
| LUAD |