
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000425456 | Fip1 | PF05182.13 | 9e-25 | 1 | 1 |
| ENSP00000302993 | Fip1 | PF05182.13 | 2.9e-24 | 1 | 1 |
| ENSP00000351383 | Fip1 | PF05182.13 | 3.4e-24 | 1 | 1 |
| ENSP00000336752 | Fip1 | PF05182.13 | 3.4e-24 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000513975 | FIP1L1-212 | 1464 | - | - | - (aa) | - | - |
| ENST00000337488 | FIP1L1-202 | 2198 | XM_005265769 | ENSP00000336752 | 594 (aa) | XP_005265826 | Q6UN15 |
| ENST00000504094 | FIP1L1-204 | 2177 | - | ENSP00000421691 | 254 (aa) | - | H0Y8P7 |
| ENST00000513008 | FIP1L1-211 | 1070 | - | - | - (aa) | - | - |
| ENST00000507206 | FIP1L1-206 | 341 | - | - | - (aa) | - | - |
| ENST00000306932 | FIP1L1-201 | 1878 | XM_005265781 | ENSP00000302993 | 520 (aa) | XP_005265838 | Q6UN15 |
| ENST00000511055 | FIP1L1-209 | 723 | - | - | - (aa) | - | - |
| ENST00000511376 | FIP1L1-210 | 582 | - | - | - (aa) | - | - |
| ENST00000510668 | FIP1L1-208 | 599 | - | - | - (aa) | - | - |
| ENST00000358575 | FIP1L1-203 | 2180 | XM_017008664 | ENSP00000351383 | 588 (aa) | XP_016864153 | Q6UN15 |
| ENST00000514543 | FIP1L1-213 | 1465 | - | ENSP00000421241 | 26 (aa) | - | H0Y8J4 |
| ENST00000507922 | FIP1L1-207 | 1335 | - | ENSP00000425456 | 378 (aa) | - | Q6UN15 |
| ENST00000505125 | FIP1L1-205 | 3180 | - | - | - (aa) | - | - |
| Pathway ID | Pathway Name | Source |
|---|---|---|
| hsa03015 | mRNA surveillance pathway | KEGG |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000145216 | Potassium | 5.5200000E-005 | - |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000145216 | rs13142904 | 4 | 53452247 | C | Macrophage Migration Inhibitory Factor levels | 27989323 | [0.14-0.31] SD units increase | 0.223 | EFO_0008221 |
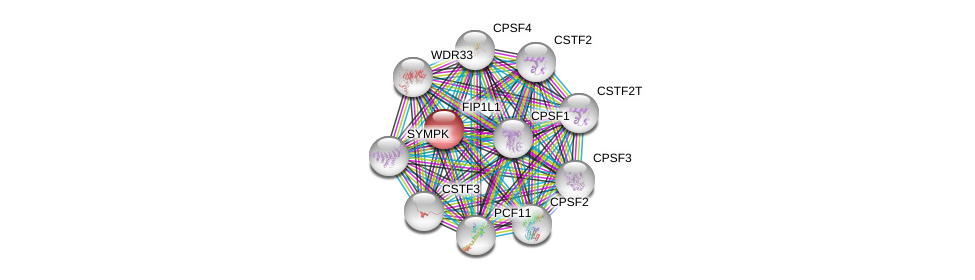
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000145216 | FIP1L1 | 100 | 100.000 | ENSANAG00000032461 | FIP1L1 | 100 | 100.000 | Aotus_nancymaae |
| ENSG00000145216 | FIP1L1 | 100 | 100.000 | ENSCJAG00000018490 | FIP1L1 | 100 | 100.000 | Callithrix_jacchus |
| ENSG00000145216 | FIP1L1 | 100 | 99.320 | ENSCAFG00020001745 | FIP1L1 | 100 | 99.320 | Canis_lupus_dingo |
| ENSG00000145216 | FIP1L1 | 100 | 97.500 | ENSTSYG00000002808 | FIP1L1 | 100 | 97.500 | Carlito_syrichta |
| ENSG00000145216 | FIP1L1 | 100 | 98.425 | ENSCSAG00000004975 | FIP1L1 | 100 | 98.342 | Chlorocebus_sabaeus |
| ENSG00000145216 | FIP1L1 | 100 | 100.000 | ENSCANG00000004785 | FIP1L1 | 100 | 100.000 | Colobus_angolensis_palliatus |
| ENSG00000145216 | FIP1L1 | 100 | 92.523 | ENSDORG00000005629 | - | 100 | 92.523 | Dipodomys_ordii |
| ENSG00000145216 | FIP1L1 | 100 | 99.663 | ENSECAG00000019415 | FIP1L1 | 100 | 99.663 | Equus_caballus |
| ENSG00000145216 | FIP1L1 | 100 | 100.000 | ENSGGOG00000006427 | FIP1L1 | 100 | 100.000 | Gorilla_gorilla |
| ENSG00000145216 | FIP1L1 | 100 | 98.822 | ENSLAFG00000002605 | FIP1L1 | 100 | 98.822 | Loxodonta_africana |
| ENSG00000145216 | FIP1L1 | 100 | 100.000 | ENSMFAG00000034699 | FIP1L1 | 100 | 100.000 | Macaca_fascicularis |
| ENSG00000145216 | FIP1L1 | 100 | 100.000 | ENSMNEG00000026332 | FIP1L1 | 100 | 100.000 | Macaca_nemestrina |
| ENSG00000145216 | FIP1L1 | 100 | 100.000 | ENSMLEG00000041763 | FIP1L1 | 100 | 100.000 | Mandrillus_leucophaeus |
| ENSG00000145216 | FIP1L1 | 99 | 99.204 | ENSMICG00000008149 | FIP1L1 | 100 | 93.077 | Microcebus_murinus |
| ENSG00000145216 | FIP1L1 | 100 | 100.000 | ENSNLEG00000008187 | FIP1L1 | 100 | 100.000 | Nomascus_leucogenys |
| ENSG00000145216 | FIP1L1 | 100 | 97.244 | ENSOCUG00000000100 | FIP1L1 | 100 | 96.186 | Oryctolagus_cuniculus |
| ENSG00000145216 | FIP1L1 | 100 | 98.824 | ENSOGAG00000009545 | FIP1L1 | 100 | 98.824 | Otolemur_garnettii |
| ENSG00000145216 | FIP1L1 | 100 | 99.327 | ENSPPRG00000002221 | FIP1L1 | 100 | 99.327 | Panthera_pardus |
| ENSG00000145216 | FIP1L1 | 100 | 97.244 | ENSPTIG00000010757 | FIP1L1 | 95 | 99.292 | Panthera_tigris_altaica |
| ENSG00000145216 | FIP1L1 | 100 | 100.000 | ENSPTRG00000016060 | FIP1L1 | 100 | 100.000 | Pan_troglodytes |
| ENSG00000145216 | FIP1L1 | 100 | 98.507 | ENSPTRG00000050184 | - | 100 | 98.507 | Pan_troglodytes |
| ENSG00000145216 | FIP1L1 | 100 | 100.000 | ENSPANG00000011213 | FIP1L1 | 100 | 100.000 | Papio_anubis |
| ENSG00000145216 | FIP1L1 | 100 | 98.425 | ENSPPYG00000014773 | FIP1L1 | 100 | 98.342 | Pongo_abelii |
| ENSG00000145216 | FIP1L1 | 99 | 99.469 | ENSPCOG00000021280 | FIP1L1 | 82 | 99.289 | Propithecus_coquereli |
| ENSG00000145216 | FIP1L1 | 100 | 100.000 | ENSRBIG00000042397 | FIP1L1 | 100 | 100.000 | Rhinopithecus_bieti |
| ENSG00000145216 | FIP1L1 | 100 | 99.660 | ENSSSCG00000031636 | FIP1L1 | 100 | 99.660 | Sus_scrofa |
| ENSG00000145216 | FIP1L1 | 99 | 93.112 | ENSTTRG00000015793 | FIP1L1 | 100 | 96.909 | Tursiops_truncatus |
| ENSG00000145216 | FIP1L1 | 99 | 99.735 | ENSUAMG00000010607 | FIP1L1 | 98 | 99.391 | Ursus_americanus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000398 | mRNA splicing, via spliceosome | - | TAS | Process |
| GO:0003723 | RNA binding | 22658674.22681889. | HDA | Function |
| GO:0003723 | RNA binding | 21873635. | IBA | Function |
| GO:0005515 | protein binding | 15937220.19224921.25416956.29276085. | IPI | Function |
| GO:0005654 | nucleoplasm | - | IDA | Component |
| GO:0005654 | nucleoplasm | - | TAS | Component |
| GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex | 21873635. | IBA | Component |
| GO:0006369 | termination of RNA polymerase II transcription | - | TAS | Process |
| GO:0006378 | mRNA polyadenylation | 21873635. | IBA | Process |
| GO:0006406 | mRNA export from nucleus | - | TAS | Process |
| GO:0031124 | mRNA 3'-end processing | - | TAS | Process |
| GO:0098789 | pre-mRNA cleavage required for polyadenylation | 21873635. | IBA | Process |