
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000376007 | CAF1 | PF04857.20 | 3.7e-79 | 1 | 1 |
| ENSP00000476448 | CAF1 | PF04857.20 | 8.7e-72 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000609334 | PNLDC1-203 | 571 | - | - | - (aa) | - | - |
| ENST00000610273 | PNLDC1-207 | 1940 | XM_011535491 | ENSP00000476448 | 520 (aa) | XP_011533793 | Q8NA58 |
| ENST00000275275 | PNLDC1-201 | 1921 | - | ENSP00000275275 | 39 (aa) | - | V9GYA0 |
| ENST00000610041 | PNLDC1-205 | 965 | - | ENSP00000476769 | 39 (aa) | - | V9GYA0 |
| ENST00000609658 | PNLDC1-204 | 475 | - | - | - (aa) | - | - |
| ENST00000610048 | PNLDC1-206 | 578 | - | ENSP00000476544 | 39 (aa) | - | V9GYA0 |
| ENST00000392167 | PNLDC1-202 | 1816 | XM_017010312 | ENSP00000376007 | 531 (aa) | XP_016865801 | Q8NA58 |
| Pathway ID | Pathway Name | Source |
|---|---|---|
| hsa03018 | RNA degradation | KEGG |
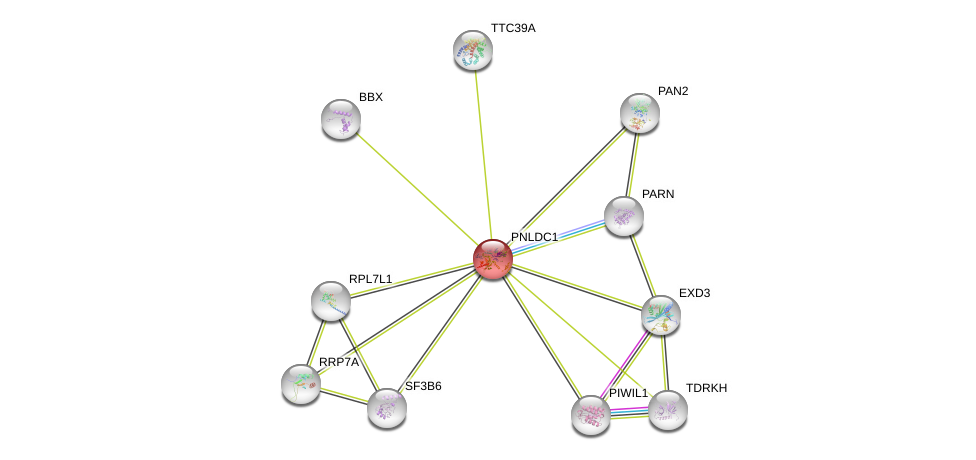
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000146453 | PNLDC1 | 99 | 86.932 | ENSAMEG00000015371 | PNLDC1 | 99 | 86.932 | Ailuropoda_melanoleuca |
| ENSG00000146453 | PNLDC1 | 99 | 58.046 | ENSAPLG00000007014 | PNLDC1 | 99 | 58.046 | Anas_platyrhynchos |
| ENSG00000146453 | PNLDC1 | 100 | 59.962 | ENSACAG00000013667 | PNLDC1 | 100 | 59.962 | Anolis_carolinensis |
| ENSG00000146453 | PNLDC1 | 83 | 30.043 | ENSACAG00000004973 | PARN | 69 | 30.043 | Anolis_carolinensis |
| ENSG00000146453 | PNLDC1 | 99 | 93.019 | ENSANAG00000034713 | PNLDC1 | 100 | 92.955 | Aotus_nancymaae |
| ENSG00000146453 | PNLDC1 | 99 | 85.851 | ENSBTAG00000015166 | PNLDC1 | 99 | 86.042 | Bos_taurus |
| ENSG00000146453 | PNLDC1 | 99 | 92.830 | ENSCJAG00000011865 | PNLDC1 | 99 | 93.083 | Callithrix_jacchus |
| ENSG00000146453 | PNLDC1 | 99 | 87.925 | ENSCAFG00000000740 | PNLDC1 | 99 | 87.925 | Canis_familiaris |
| ENSG00000146453 | PNLDC1 | 99 | 86.064 | ENSCAFG00020017742 | PNLDC1 | 99 | 86.064 | Canis_lupus_dingo |
| ENSG00000146453 | PNLDC1 | 99 | 85.472 | ENSCHIG00000003141 | PNLDC1 | 99 | 85.472 | Capra_hircus |
| ENSG00000146453 | PNLDC1 | 99 | 90.377 | ENSTSYG00000005606 | PNLDC1 | 99 | 90.377 | Carlito_syrichta |
| ENSG00000146453 | PNLDC1 | 99 | 61.509 | ENSCAPG00000011893 | PNLDC1 | 99 | 61.509 | Cavia_aperea |
| ENSG00000146453 | PNLDC1 | 99 | 78.719 | ENSCPOG00000019565 | PNLDC1 | 99 | 78.719 | Cavia_porcellus |
| ENSG00000146453 | PNLDC1 | 99 | 94.151 | ENSCCAG00000026232 | PNLDC1 | 99 | 94.151 | Cebus_capucinus |
| ENSG00000146453 | PNLDC1 | 99 | 96.415 | ENSCATG00000033196 | PNLDC1 | 99 | 96.415 | Cercocebus_atys |
| ENSG00000146453 | PNLDC1 | 99 | 83.962 | ENSCLAG00000003228 | PNLDC1 | 99 | 83.962 | Chinchilla_lanigera |
| ENSG00000146453 | PNLDC1 | 99 | 95.393 | ENSCSAG00000012478 | PNLDC1 | 99 | 95.393 | Chlorocebus_sabaeus |
| ENSG00000146453 | PNLDC1 | 96 | 65.430 | ENSCPBG00000004050 | PNLDC1 | 100 | 65.430 | Chrysemys_picta_bellii |
| ENSG00000146453 | PNLDC1 | 99 | 95.183 | ENSCANG00000043076 | PNLDC1 | 99 | 95.183 | Colobus_angolensis_palliatus |
| ENSG00000146453 | PNLDC1 | 99 | 66.038 | ENSCGRG00001011981 | - | 99 | 66.038 | Cricetulus_griseus_chok1gshd |
| ENSG00000146453 | PNLDC1 | 99 | 81.321 | ENSCGRG00001007470 | - | 99 | 81.321 | Cricetulus_griseus_chok1gshd |
| ENSG00000146453 | PNLDC1 | 99 | 81.321 | ENSCGRG00000010221 | - | 99 | 81.321 | Cricetulus_griseus_crigri |
| ENSG00000146453 | PNLDC1 | 99 | 68.113 | ENSCGRG00000003103 | - | 99 | 68.113 | Cricetulus_griseus_crigri |
| ENSG00000146453 | PNLDC1 | 99 | 86.174 | ENSDNOG00000013709 | PNLDC1 | 99 | 86.174 | Dasypus_novemcinctus |
| ENSG00000146453 | PNLDC1 | 99 | 79.550 | ENSDORG00000011517 | Pnldc1 | 99 | 86.901 | Dipodomys_ordii |
| ENSG00000146453 | PNLDC1 | 93 | 76.543 | ENSETEG00000007587 | PNLDC1 | 98 | 76.543 | Echinops_telfairi |
| ENSG00000146453 | PNLDC1 | 99 | 79.926 | ENSEASG00005019029 | PNLDC1 | 99 | 79.926 | Equus_asinus_asinus |
| ENSG00000146453 | PNLDC1 | 99 | 88.302 | ENSECAG00000012898 | PNLDC1 | 99 | 88.302 | Equus_caballus |
| ENSG00000146453 | PNLDC1 | 69 | 81.250 | ENSEEUG00000005717 | - | 86 | 81.250 | Erinaceus_europaeus |
| ENSG00000146453 | PNLDC1 | 99 | 88.068 | ENSFCAG00000002848 | PNLDC1 | 99 | 88.068 | Felis_catus |
| ENSG00000146453 | PNLDC1 | 96 | 57.455 | ENSFALG00000012081 | PNLDC1 | 99 | 57.455 | Ficedula_albicollis |
| ENSG00000146453 | PNLDC1 | 99 | 84.499 | ENSFDAG00000021334 | PNLDC1 | 99 | 84.499 | Fukomys_damarensis |
| ENSG00000146453 | PNLDC1 | 79 | 58.670 | ENSGALG00000011624 | PNLDC1 | 82 | 58.670 | Gallus_gallus |
| ENSG00000146453 | PNLDC1 | 99 | 66.408 | ENSGAGG00000015276 | PNLDC1 | 100 | 67.355 | Gopherus_agassizii |
| ENSG00000146453 | PNLDC1 | 99 | 99.245 | ENSGGOG00000014077 | PNLDC1 | 99 | 99.245 | Gorilla_gorilla |
| ENSG00000146453 | PNLDC1 | 99 | 84.906 | ENSHGLG00000006889 | PNLDC1 | 99 | 84.906 | Heterocephalus_glaber_female |
| ENSG00000146453 | PNLDC1 | 99 | 84.906 | ENSHGLG00100018275 | PNLDC1 | 99 | 84.906 | Heterocephalus_glaber_male |
| ENSG00000146453 | PNLDC1 | 99 | 88.658 | ENSSTOG00000005595 | PNLDC1 | 99 | 88.658 | Ictidomys_tridecemlineatus |
| ENSG00000146453 | PNLDC1 | 99 | 80.266 | ENSJJAG00000009937 | Pnldc1 | 99 | 80.266 | Jaculus_jaculus |
| ENSG00000146453 | PNLDC1 | 59 | 85.397 | ENSLAFG00000030189 | PNLDC1 | 92 | 85.397 | Loxodonta_africana |
| ENSG00000146453 | PNLDC1 | 99 | 96.415 | ENSMFAG00000041296 | PNLDC1 | 99 | 96.415 | Macaca_fascicularis |
| ENSG00000146453 | PNLDC1 | 99 | 96.415 | ENSMMUG00000004181 | PNLDC1 | 99 | 96.415 | Macaca_mulatta |
| ENSG00000146453 | PNLDC1 | 99 | 96.226 | ENSMNEG00000034271 | PNLDC1 | 99 | 96.226 | Macaca_nemestrina |
| ENSG00000146453 | PNLDC1 | 99 | 92.830 | ENSMLEG00000044059 | PNLDC1 | 99 | 92.830 | Mandrillus_leucophaeus |
| ENSG00000146453 | PNLDC1 | 98 | 58.743 | ENSMGAG00000012470 | PNLDC1 | 97 | 58.743 | Meleagris_gallopavo |
| ENSG00000146453 | PNLDC1 | 99 | 66.917 | ENSMAUG00000012298 | Pnldc1 | 100 | 66.917 | Mesocricetus_auratus |
| ENSG00000146453 | PNLDC1 | 99 | 88.491 | ENSMICG00000008021 | PNLDC1 | 99 | 88.491 | Microcebus_murinus |
| ENSG00000146453 | PNLDC1 | 99 | 85.902 | ENSMOCG00000014287 | Pnldc1 | 99 | 85.902 | Microtus_ochrogaster |
| ENSG00000146453 | PNLDC1 | 99 | 75.614 | ENSMODG00000007420 | PNLDC1 | 99 | 75.614 | Monodelphis_domestica |
| ENSG00000146453 | PNLDC1 | 99 | 85.985 | MGP_CAROLIEiJ_G0021045 | Pnldc1 | 99 | 85.985 | Mus_caroli |
| ENSG00000146453 | PNLDC1 | 99 | 85.985 | ENSMUSG00000073460 | Pnldc1 | 100 | 88.571 | Mus_musculus |
| ENSG00000146453 | PNLDC1 | 99 | 86.200 | MGP_PahariEiJ_G0023361 | Pnldc1 | 99 | 86.200 | Mus_pahari |
| ENSG00000146453 | PNLDC1 | 99 | 85.795 | MGP_SPRETEiJ_G0021975 | Pnldc1 | 99 | 85.795 | Mus_spretus |
| ENSG00000146453 | PNLDC1 | 99 | 86.792 | ENSMPUG00000004920 | PNLDC1 | 99 | 86.792 | Mustela_putorius_furo |
| ENSG00000146453 | PNLDC1 | 99 | 86.415 | ENSMLUG00000000747 | PNLDC1 | 99 | 86.415 | Myotis_lucifugus |
| ENSG00000146453 | PNLDC1 | 99 | 87.358 | ENSNGAG00000017714 | Pnldc1 | 99 | 87.358 | Nannospalax_galili |
| ENSG00000146453 | PNLDC1 | 99 | 96.532 | ENSNLEG00000011282 | PNLDC1 | 99 | 96.532 | Nomascus_leucogenys |
| ENSG00000146453 | PNLDC1 | 55 | 66.667 | ENSMEUG00000014042 | - | 100 | 66.667 | Notamacropus_eugenii |
| ENSG00000146453 | PNLDC1 | 62 | 63.438 | ENSMEUG00000011926 | - | 64 | 63.438 | Notamacropus_eugenii |
| ENSG00000146453 | PNLDC1 | 79 | 86.364 | ENSOPRG00000003813 | PNLDC1 | 79 | 86.364 | Ochotona_princeps |
| ENSG00000146453 | PNLDC1 | 99 | 83.932 | ENSODEG00000008047 | PNLDC1 | 99 | 83.932 | Octodon_degus |
| ENSG00000146453 | PNLDC1 | 99 | 57.170 | ENSOANG00000012199 | PNLDC1 | 99 | 59.434 | Ornithorhynchus_anatinus |
| ENSG00000146453 | PNLDC1 | 99 | 86.742 | ENSOCUG00000013867 | PNLDC1 | 99 | 86.742 | Oryctolagus_cuniculus |
| ENSG00000146453 | PNLDC1 | 99 | 89.603 | ENSOGAG00000007331 | PNLDC1 | 100 | 89.603 | Otolemur_garnettii |
| ENSG00000146453 | PNLDC1 | 99 | 85.660 | ENSOARG00000004585 | PNLDC1 | 99 | 85.660 | Ovis_aries |
| ENSG00000146453 | PNLDC1 | 99 | 98.868 | ENSPPAG00000026651 | PNLDC1 | 99 | 98.868 | Pan_paniscus |
| ENSG00000146453 | PNLDC1 | 99 | 88.068 | ENSPPRG00000012854 | PNLDC1 | 99 | 88.068 | Panthera_pardus |
| ENSG00000146453 | PNLDC1 | 97 | 86.056 | ENSPTIG00000011501 | PNLDC1 | 99 | 83.619 | Panthera_tigris_altaica |
| ENSG00000146453 | PNLDC1 | 99 | 98.868 | ENSPTRG00000018762 | PNLDC1 | 99 | 98.868 | Pan_troglodytes |
| ENSG00000146453 | PNLDC1 | 99 | 96.604 | ENSPANG00000022009 | PNLDC1 | 99 | 96.604 | Papio_anubis |
| ENSG00000146453 | PNLDC1 | 62 | 56.000 | ENSPSIG00000011419 | PNLDC1 | 99 | 56.000 | Pelodiscus_sinensis |
| ENSG00000146453 | PNLDC1 | 99 | 86.553 | ENSPEMG00000016301 | Pnldc1 | 99 | 86.553 | Peromyscus_maniculatus_bairdii |
| ENSG00000146453 | PNLDC1 | 86 | 30.629 | ENSPMAG00000003956 | parn | 90 | 30.832 | Petromyzon_marinus |
| ENSG00000146453 | PNLDC1 | 84 | 76.538 | ENSPCIG00000023412 | PNLDC1 | 100 | 76.538 | Phascolarctos_cinereus |
| ENSG00000146453 | PNLDC1 | 99 | 90.584 | ENSPPYG00000017148 | PNLDC1 | 99 | 90.584 | Pongo_abelii |
| ENSG00000146453 | PNLDC1 | 99 | 76.705 | ENSPCAG00000014169 | PNLDC1 | 99 | 76.705 | Procavia_capensis |
| ENSG00000146453 | PNLDC1 | 99 | 89.245 | ENSPCOG00000025722 | PNLDC1 | 99 | 89.245 | Propithecus_coquereli |
| ENSG00000146453 | PNLDC1 | 73 | 89.870 | ENSPVAG00000005325 | PNLDC1 | 73 | 89.870 | Pteropus_vampyrus |
| ENSG00000146453 | PNLDC1 | 99 | 85.985 | ENSRNOG00000023140 | Pnldc1 | 100 | 86.250 | Rattus_norvegicus |
| ENSG00000146453 | PNLDC1 | 99 | 96.415 | ENSRBIG00000033806 | PNLDC1 | 99 | 96.415 | Rhinopithecus_bieti |
| ENSG00000146453 | PNLDC1 | 99 | 95.761 | ENSRROG00000043919 | PNLDC1 | 99 | 95.761 | Rhinopithecus_roxellana |
| ENSG00000146453 | PNLDC1 | 99 | 94.151 | ENSSBOG00000019135 | PNLDC1 | 99 | 94.151 | Saimiri_boliviensis_boliviensis |
| ENSG00000146453 | PNLDC1 | 99 | 76.163 | ENSSHAG00000015192 | PNLDC1 | 99 | 76.163 | Sarcophilus_harrisii |
| ENSG00000146453 | PNLDC1 | 55 | 65.398 | ENSSPUG00000004627 | PNLDC1 | 99 | 66.071 | Sphenodon_punctatus |
| ENSG00000146453 | PNLDC1 | 98 | 86.444 | ENSSSCG00000004046 | PNLDC1 | 99 | 86.444 | Sus_scrofa |
| ENSG00000146453 | PNLDC1 | 93 | 58.557 | ENSTGUG00000011418 | PNLDC1 | 99 | 58.557 | Taeniopygia_guttata |
| ENSG00000146453 | PNLDC1 | 99 | 77.043 | ENSTBEG00000009131 | PNLDC1 | 99 | 77.043 | Tupaia_belangeri |
| ENSG00000146453 | PNLDC1 | 73 | 83.117 | ENSTTRG00000010409 | PNLDC1 | 72 | 83.117 | Tursiops_truncatus |
| ENSG00000146453 | PNLDC1 | 99 | 75.000 | ENSUAMG00000015783 | PNLDC1 | 99 | 75.000 | Ursus_americanus |
| ENSG00000146453 | PNLDC1 | 97 | 80.196 | ENSUMAG00000011903 | PNLDC1 | 99 | 80.196 | Ursus_maritimus |
| ENSG00000146453 | PNLDC1 | 96 | 88.650 | ENSVVUG00000006586 | PNLDC1 | 99 | 87.860 | Vulpes_vulpes |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000175 | 3'-5'-exoribonuclease activity | 21873635. | IBA | Function |
| GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | - | IEA | Process |
| GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening | 21873635. | IBA | Process |
| GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening | 27515512. | IDA | Process |
| GO:0001825 | blastocyst formation | - | IEA | Process |
| GO:0003723 | RNA binding | - | IEA | Function |
| GO:0004535 | poly(A)-specific ribonuclease activity | 21873635. | IBA | Function |
| GO:0004535 | poly(A)-specific ribonuclease activity | 27515512. | IDA | Function |
| GO:0005737 | cytoplasm | 21873635. | IBA | Component |
| GO:0005783 | endoplasmic reticulum | 21873635. | IBA | Component |
| GO:0005783 | endoplasmic reticulum | 27515512. | IDA | Component |
| GO:0005789 | endoplasmic reticulum membrane | - | IEA | Component |
| GO:0016021 | integral component of membrane | - | IEA | Component |
| GO:0046872 | metal ion binding | - | IEA | Function |
| GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic | - | IEA | Process |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CHOL | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRP | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| THCA | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| BLCA | |||
| CESC | |||
| KIRC | |||
| KIRP | |||
| LGG | |||
| MESO | |||
| PAAD | |||
| SKCM | |||
| TGCT | |||
| THYM |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| MESO | ||
| HNSC | ||
| SKCM | ||
| BRCA | ||
| COAD | ||
| UCEC | ||
| GBM |