
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000489129 | zf-C2H2_jaz | PF12171.8 | 2e-07 | 1 | 1 |
| ENSP00000362029 | zf-C2H2_jaz | PF12171.8 | 7.2e-07 | 1 | 1 |
| ENSP00000489425 | zf-C2H2_jaz | PF12171.8 | 7.2e-07 | 1 | 1 |
| ENSP00000439244 | zf-C2H2_jaz | PF12171.8 | 9.2e-07 | 1 | 1 |
| ENSP00000486816 | zf-C2H2_jaz | PF12171.8 | 1.8e-06 | 1 | 1 |
| ENSP00000362045 | zf-C2H2_jaz | PF12171.8 | 2e-06 | 1 | 1 |
| ENSP00000398011 | zf-C2H2_jaz | PF12171.8 | 2e-06 | 1 | 1 |
| ENSP00000377232 | zf-C2H2_jaz | PF12171.8 | 2.2e-06 | 1 | 1 |
| ENSP00000362039 | zf-C2H2_jaz | PF12171.8 | 2.2e-06 | 1 | 1 |
| ENSP00000277465 | zf-C2H2_jaz | PF12171.8 | 2.5e-06 | 1 | 1 |
| ENSP00000350169 | zf-C2H2_jaz | PF12171.8 | 2.5e-06 | 1 | 1 |
| PID | Title | Method | Time | Author | Doi |
|---|---|---|---|---|---|
| 30607034 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000467178 | CIZ1-211 | 904 | - | - | - (aa) | - | - |
| ENST00000476727 | CIZ1-218 | 1205 | - | - | - (aa) | - | - |
| ENST00000485001 | CIZ1-219 | 596 | - | - | - (aa) | - | - |
| ENST00000634901 | CIZ1-228 | 3462 | - | ENSP00000489425 | 898 (aa) | - | Q9ULV3 |
| ENST00000324544 | CIZ1-202 | 809 | - | ENSP00000321780 | 252 (aa) | - | F6VD24 |
| ENST00000491487 | CIZ1-222 | 339 | - | - | - (aa) | - | - |
| ENST00000277465 | CIZ1-201 | 2742 | - | ENSP00000277465 | 870 (aa) | - | Q5SYW2 |
| ENST00000471773 | CIZ1-212 | 717 | - | - | - (aa) | - | - |
| ENST00000420484 | CIZ1-209 | 684 | - | ENSP00000407265 | 168 (aa) | - | F6WSM2 |
| ENST00000372948 | CIZ1-205 | 2858 | XM_005251891 | ENSP00000362039 | 842 (aa) | XP_005251948 | Q9ULV3 |
| ENST00000471839 | CIZ1-213 | 928 | - | ENSP00000489129 | 310 (aa) | - | A0A0U1RQR2 |
| ENST00000485862 | CIZ1-220 | 843 | - | - | - (aa) | - | - |
| ENST00000476541 | CIZ1-217 | 513 | - | - | - (aa) | - | - |
| ENST00000475471 | CIZ1-215 | 836 | - | - | - (aa) | - | - |
| ENST00000629610 | CIZ1-226 | 2855 | - | ENSP00000486816 | 797 (aa) | - | Q9ULV3 |
| ENST00000498156 | CIZ1-224 | 642 | - | - | - (aa) | - | - |
| ENST00000357558 | CIZ1-203 | 2907 | - | ENSP00000350169 | 870 (aa) | - | Q5SYW2 |
| ENST00000393608 | CIZ1-207 | 2645 | - | ENSP00000377232 | 837 (aa) | - | Q9BTG3 |
| ENST00000461765 | CIZ1-210 | 677 | - | - | - (aa) | - | - |
| ENST00000634501 | CIZ1-227 | 1996 | - | - | - (aa) | - | - |
| ENST00000538431 | CIZ1-225 | 2996 | XM_017014594 | ENSP00000439244 | 954 (aa) | XP_016870083 | F5H2X7 |
| ENST00000476239 | CIZ1-216 | 853 | - | - | - (aa) | - | - |
| ENST00000372938 | CIZ1-204 | 2984 | XM_005251888 | ENSP00000362029 | 898 (aa) | XP_005251945 | Q9ULV3 |
| ENST00000491954 | CIZ1-223 | 955 | - | - | - (aa) | - | - |
| ENST00000415526 | CIZ1-208 | 2508 | - | ENSP00000398011 | 820 (aa) | - | H0Y5D5 |
| ENST00000372954 | CIZ1-206 | 2731 | XM_005251893 | ENSP00000362045 | 818 (aa) | XP_005251950 | Q9ULV3 |
| ENST00000488559 | CIZ1-221 | 952 | - | - | - (aa) | - | - |
| ENST00000474442 | CIZ1-214 | 457 | - | - | - (aa) | - | - |
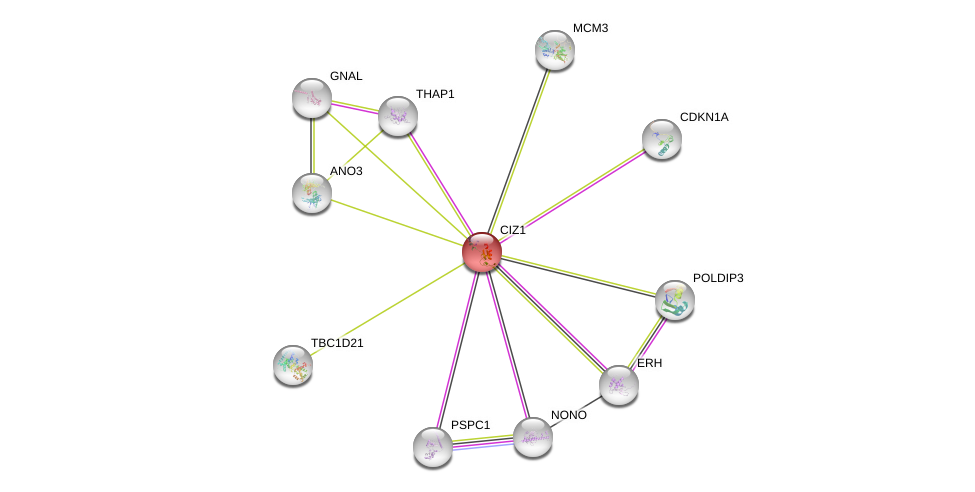
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000148337 | CIZ1 | 99 | 83.495 | ENSAMEG00000015276 | CIZ1 | 98 | 75.669 | Ailuropoda_melanoleuca |
| ENSG00000148337 | CIZ1 | 99 | 90.968 | ENSANAG00000037943 | CIZ1 | 98 | 87.931 | Aotus_nancymaae |
| ENSG00000148337 | CIZ1 | 85 | 36.364 | ENSACLG00000008591 | - | 69 | 37.539 | Astatotilapia_calliptera |
| ENSG00000148337 | CIZ1 | 99 | 83.117 | ENSBTAG00000012274 | CIZ1 | 98 | 77.941 | Bos_taurus |
| ENSG00000148337 | CIZ1 | 99 | 93.528 | ENSCJAG00000019948 | CIZ1 | 100 | 85.639 | Callithrix_jacchus |
| ENSG00000148337 | CIZ1 | 99 | 84.416 | ENSCAFG00000020086 | CIZ1 | 99 | 77.918 | Canis_familiaris |
| ENSG00000148337 | CIZ1 | 99 | 84.416 | ENSCAFG00020016118 | CIZ1 | 99 | 77.198 | Canis_lupus_dingo |
| ENSG00000148337 | CIZ1 | 99 | 84.740 | ENSCHIG00000006845 | CIZ1 | 97 | 76.289 | Capra_hircus |
| ENSG00000148337 | CIZ1 | 99 | 87.702 | ENSTSYG00000031123 | CIZ1 | 96 | 81.273 | Carlito_syrichta |
| ENSG00000148337 | CIZ1 | 99 | 73.482 | ENSCAPG00000016903 | CIZ1 | 96 | 67.268 | Cavia_aperea |
| ENSG00000148337 | CIZ1 | 99 | 74.121 | ENSCPOG00000006863 | CIZ1 | 94 | 70.305 | Cavia_porcellus |
| ENSG00000148337 | CIZ1 | 99 | 90.968 | ENSCCAG00000026440 | CIZ1 | 95 | 90.769 | Cebus_capucinus |
| ENSG00000148337 | CIZ1 | 99 | 98.058 | ENSCATG00000034742 | CIZ1 | 99 | 95.818 | Cercocebus_atys |
| ENSG00000148337 | CIZ1 | 99 | 69.876 | ENSCLAG00000001584 | CIZ1 | 93 | 71.469 | Chinchilla_lanigera |
| ENSG00000148337 | CIZ1 | 95 | 95.250 | ENSCSAG00000014060 | CIZ1 | 99 | 95.695 | Chlorocebus_sabaeus |
| ENSG00000148337 | CIZ1 | 99 | 98.058 | ENSCANG00000040660 | CIZ1 | 99 | 96.556 | Colobus_angolensis_palliatus |
| ENSG00000148337 | CIZ1 | 99 | 77.273 | ENSCGRG00001014777 | Ciz1 | 95 | 73.159 | Cricetulus_griseus_chok1gshd |
| ENSG00000148337 | CIZ1 | 89 | 44.043 | ENSDARG00000089461 | ciz1b | 56 | 43.466 | Danio_rerio |
| ENSG00000148337 | CIZ1 | 99 | 82.524 | ENSDNOG00000036104 | CIZ1 | 97 | 78.385 | Dasypus_novemcinctus |
| ENSG00000148337 | CIZ1 | 96 | 74.161 | ENSDORG00000012341 | Ciz1 | 98 | 63.573 | Dipodomys_ordii |
| ENSG00000148337 | CIZ1 | 99 | 70.968 | ENSETEG00000005455 | CIZ1 | 100 | 67.707 | Echinops_telfairi |
| ENSG00000148337 | CIZ1 | 98 | 73.300 | ENSEASG00005020348 | CIZ1 | 100 | 73.538 | Equus_asinus_asinus |
| ENSG00000148337 | CIZ1 | 99 | 83.172 | ENSECAG00000007116 | CIZ1 | 95 | 80.456 | Equus_caballus |
| ENSG00000148337 | CIZ1 | 71 | 86.667 | ENSEEUG00000012590 | - | 68 | 78.873 | Erinaceus_europaeus |
| ENSG00000148337 | CIZ1 | 91 | 41.924 | ENSELUG00000023867 | - | 78 | 44.898 | Esox_lucius |
| ENSG00000148337 | CIZ1 | 99 | 83.226 | ENSFCAG00000014677 | CIZ1 | 96 | 79.753 | Felis_catus |
| ENSG00000148337 | CIZ1 | 99 | 72.500 | ENSFDAG00000016651 | CIZ1 | 98 | 70.246 | Fukomys_damarensis |
| ENSG00000148337 | CIZ1 | 85 | 36.502 | ENSFHEG00000009254 | - | 63 | 37.987 | Fundulus_heteroclitus |
| ENSG00000148337 | CIZ1 | 87 | 37.895 | ENSGMOG00000018734 | - | 99 | 38.686 | Gadus_morhua |
| ENSG00000148337 | CIZ1 | 56 | 53.409 | ENSGALG00000042325 | - | 54 | 57.843 | Gallus_gallus |
| ENSG00000148337 | CIZ1 | 86 | 36.594 | ENSGACG00000016651 | - | 56 | 40.239 | Gasterosteus_aculeatus |
| ENSG00000148337 | CIZ1 | 99 | 99.353 | ENSGGOG00000014999 | CIZ1 | 100 | 97.691 | Gorilla_gorilla |
| ENSG00000148337 | CIZ1 | 85 | 36.364 | ENSHBUG00000021945 | - | 69 | 37.855 | Haplochromis_burtoni |
| ENSG00000148337 | CIZ1 | 99 | 71.246 | ENSHGLG00000011900 | CIZ1 | 97 | 72.738 | Heterocephalus_glaber_female |
| ENSG00000148337 | CIZ1 | 99 | 71.246 | ENSHGLG00100018035 | CIZ1 | 97 | 72.738 | Heterocephalus_glaber_male |
| ENSG00000148337 | CIZ1 | 85 | 80.970 | ENSSTOG00000033639 | - | 99 | 88.353 | Ictidomys_tridecemlineatus |
| ENSG00000148337 | CIZ1 | 81 | 73.016 | ENSJJAG00000015415 | - | 100 | 77.165 | Jaculus_jaculus |
| ENSG00000148337 | CIZ1 | 86 | 35.185 | ENSKMAG00000006597 | - | 68 | 37.097 | Kryptolebias_marmoratus |
| ENSG00000148337 | CIZ1 | 87 | 32.990 | ENSLBEG00000007927 | - | 67 | 37.654 | Labrus_bergylta |
| ENSG00000148337 | CIZ1 | 99 | 77.922 | ENSLAFG00000005106 | CIZ1 | 95 | 76.208 | Loxodonta_africana |
| ENSG00000148337 | CIZ1 | 99 | 97.735 | ENSMFAG00000034538 | CIZ1 | 99 | 95.941 | Macaca_fascicularis |
| ENSG00000148337 | CIZ1 | 99 | 97.411 | ENSMMUG00000013712 | CIZ1 | 99 | 95.699 | Macaca_mulatta |
| ENSG00000148337 | CIZ1 | 99 | 98.058 | ENSMNEG00000041837 | CIZ1 | 99 | 95.941 | Macaca_nemestrina |
| ENSG00000148337 | CIZ1 | 99 | 97.735 | ENSMLEG00000035347 | CIZ1 | 100 | 95.111 | Mandrillus_leucophaeus |
| ENSG00000148337 | CIZ1 | 86 | 36.162 | ENSMAMG00000019077 | - | 72 | 39.041 | Mastacembelus_armatus |
| ENSG00000148337 | CIZ1 | 85 | 36.364 | ENSMZEG00005000299 | - | 69 | 37.539 | Maylandia_zebra |
| ENSG00000148337 | CIZ1 | 56 | 53.409 | ENSMGAG00000004067 | - | 90 | 58.416 | Meleagris_gallopavo |
| ENSG00000148337 | CIZ1 | 81 | 65.354 | ENSMAUG00000015443 | - | 99 | 72.458 | Mesocricetus_auratus |
| ENSG00000148337 | CIZ1 | 99 | 87.662 | ENSMICG00000003411 | CIZ1 | 86 | 80.319 | Microcebus_murinus |
| ENSG00000148337 | CIZ1 | 79 | 76.016 | ENSMOCG00000009560 | - | 99 | 83.043 | Microtus_ochrogaster |
| ENSG00000148337 | CIZ1 | 98 | 72.876 | MGP_CAROLIEiJ_G0023430 | Ciz1 | 99 | 77.682 | Mus_caroli |
| ENSG00000148337 | CIZ1 | 98 | 72.403 | ENSMUSG00000039205 | Ciz1 | 99 | 76.596 | Mus_musculus |
| ENSG00000148337 | CIZ1 | 99 | 71.795 | MGP_PahariEiJ_G0024887 | Ciz1 | 99 | 77.215 | Mus_pahari |
| ENSG00000148337 | CIZ1 | 98 | 71.661 | MGP_SPRETEiJ_G0024347 | Ciz1 | 99 | 75.745 | Mus_spretus |
| ENSG00000148337 | CIZ1 | 99 | 82.848 | ENSMPUG00000013449 | CIZ1 | 97 | 77.047 | Mustela_putorius_furo |
| ENSG00000148337 | CIZ1 | 99 | 75.325 | ENSMLUG00000016220 | CIZ1 | 94 | 70.433 | Myotis_lucifugus |
| ENSG00000148337 | CIZ1 | 79 | 73.577 | ENSNGAG00000016410 | - | 99 | 80.000 | Nannospalax_galili |
| ENSG00000148337 | CIZ1 | 90 | 78.182 | ENSOPRG00000016419 | CIZ1 | 96 | 62.471 | Ochotona_princeps |
| ENSG00000148337 | CIZ1 | 98 | 68.440 | ENSODEG00000017392 | CIZ1 | 97 | 69.258 | Octodon_degus |
| ENSG00000148337 | CIZ1 | 99 | 71.104 | ENSOCUG00000015985 | CIZ1 | 98 | 70.759 | Oryctolagus_cuniculus |
| ENSG00000148337 | CIZ1 | 85 | 36.604 | ENSORLG00000027096 | - | 68 | 38.411 | Oryzias_latipes |
| ENSG00000148337 | CIZ1 | 84 | 33.333 | ENSORLG00015000909 | - | 79 | 36.755 | Oryzias_latipes_hsok |
| ENSG00000148337 | CIZ1 | 99 | 78.457 | ENSOGAG00000013198 | CIZ1 | 100 | 75.651 | Otolemur_garnettii |
| ENSG00000148337 | CIZ1 | 99 | 82.468 | ENSOARG00000010311 | CIZ1 | 97 | 75.032 | Ovis_aries |
| ENSG00000148337 | CIZ1 | 100 | 99.405 | ENSPPAG00000030730 | CIZ1 | 100 | 99.389 | Pan_paniscus |
| ENSG00000148337 | CIZ1 | 99 | 83.819 | ENSPPRG00000012431 | CIZ1 | 98 | 79.393 | Panthera_pardus |
| ENSG00000148337 | CIZ1 | 99 | 83.495 | ENSPTIG00000009492 | CIZ1 | 97 | 75.162 | Panthera_tigris_altaica |
| ENSG00000148337 | CIZ1 | 99 | 100.000 | ENSPTRG00000051341 | - | 100 | 98.957 | Pan_troglodytes |
| ENSG00000148337 | CIZ1 | 99 | 100.000 | ENSPTRG00000045021 | - | 100 | 96.346 | Pan_troglodytes |
| ENSG00000148337 | CIZ1 | 99 | 97.735 | ENSPANG00000024871 | CIZ1 | 100 | 95.111 | Papio_anubis |
| ENSG00000148337 | CIZ1 | 96 | 77.441 | ENSPEMG00000002769 | Ciz1 | 100 | 80.932 | Peromyscus_maniculatus_bairdii |
| ENSG00000148337 | CIZ1 | 85 | 35.484 | ENSPREG00000003135 | - | 65 | 40.830 | Poecilia_reticulata |
| ENSG00000148337 | CIZ1 | 100 | 96.887 | ENSPPYG00000019645 | CIZ1 | 100 | 95.868 | Pongo_abelii |
| ENSG00000148337 | CIZ1 | 99 | 74.351 | ENSPCAG00000011814 | CIZ1 | 95 | 73.430 | Procavia_capensis |
| ENSG00000148337 | CIZ1 | 99 | 84.691 | ENSPCOG00000027303 | CIZ1 | 97 | 81.594 | Propithecus_coquereli |
| ENSG00000148337 | CIZ1 | 99 | 80.583 | ENSPVAG00000002437 | CIZ1 | 96 | 75.361 | Pteropus_vampyrus |
| ENSG00000148337 | CIZ1 | 99 | 73.871 | ENSRNOG00000013442 | Ciz1 | 95 | 68.486 | Rattus_norvegicus |
| ENSG00000148337 | CIZ1 | 99 | 98.382 | ENSRBIG00000034423 | CIZ1 | 99 | 96.556 | Rhinopithecus_bieti |
| ENSG00000148337 | CIZ1 | 99 | 98.058 | ENSRROG00000037746 | CIZ1 | 99 | 96.433 | Rhinopithecus_roxellana |
| ENSG00000148337 | CIZ1 | 100 | 90.241 | ENSSBOG00000030332 | CIZ1 | 95 | 91.656 | Saimiri_boliviensis_boliviensis |
| ENSG00000148337 | CIZ1 | 88 | 56.786 | ENSSHAG00000018645 | - | 83 | 59.627 | Sarcophilus_harrisii |
| ENSG00000148337 | CIZ1 | 76 | 72.464 | ENSSARG00000002139 | - | 78 | 71.875 | Sorex_araneus |
| ENSG00000148337 | CIZ1 | 98 | 85.809 | ENSSSCG00000005640 | CIZ1 | 97 | 78.374 | Sus_scrofa |
| ENSG00000148337 | CIZ1 | 89 | 53.979 | ENSTGUG00000003828 | - | 59 | 58.491 | Taeniopygia_guttata |
| ENSG00000148337 | CIZ1 | 97 | 76.000 | ENSTBEG00000014306 | CIZ1 | 99 | 70.078 | Tupaia_belangeri |
| ENSG00000148337 | CIZ1 | 99 | 85.714 | ENSTTRG00000016480 | CIZ1 | 100 | 80.046 | Tursiops_truncatus |
| ENSG00000148337 | CIZ1 | 99 | 84.142 | ENSUAMG00000009953 | CIZ1 | 100 | 78.261 | Ursus_americanus |
| ENSG00000148337 | CIZ1 | 95 | 84.122 | ENSUMAG00000022712 | - | 94 | 75.622 | Ursus_maritimus |
| ENSG00000148337 | CIZ1 | 55 | 95.522 | ENSVPAG00000009256 | - | 77 | 94.444 | Vicugna_pacos |
| ENSG00000148337 | CIZ1 | 99 | 85.390 | ENSVVUG00000013406 | CIZ1 | 91 | 77.751 | Vulpes_vulpes |
| ENSG00000148337 | CIZ1 | 87 | 35.688 | ENSXMAG00000028127 | - | 64 | 39.446 | Xiphophorus_maculatus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0003676 | nucleic acid binding | - | IEA | Function |
| GO:0005515 | protein binding | 18654987. | IPI | Function |
| GO:0005634 | nucleus | 10529385. | TAS | Component |
| GO:0005654 | nucleoplasm | - | IDA | Component |
| GO:0005886 | plasma membrane | - | IDA | Component |
| GO:0008270 | zinc ion binding | - | IEA | Function |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| KIRP | |||||||
| KIRP | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| READ | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| TGCT | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| CHOL | |||
| LAML | |||
| LGG | |||
| LIHC | |||
| PRAD | |||
| THCA | |||
| THYM | |||
| UCS |
| Cancer | P-value | Q-value |
|---|---|---|
| SARC | ||
| MESO | ||
| ACC | ||
| UCS | ||
| SKCM | ||
| PRAD | ||
| KIRP | ||
| COAD | ||
| PAAD | ||
| PCPG | ||
| BLCA | ||
| LIHC | ||
| LGG | ||
| UVM | ||
| OV |