
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000278505 | Endonuclease_NS | PF01223.23 | 2.1e-05 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000278505 | ENDOD1-201 | 4687 | - | ENSP00000278505 | 500 (aa) | - | O94919 |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000149218 | Job Satisfaction | 3.1110000E-005 | - |
| ENSG00000149218 | Lipoprotein(a) | 9.7580000E-005 | - |
| ENSG00000149218 | Platelet Function Tests | 7.9200000E-010 | - |
| ENSG00000149218 | Platelet Function Tests | 5.2600000E-006 | - |
| ENSG00000149218 | Platelet Function Tests | 6.4400000E-006 | - |
| ENSG00000149218 | Platelet Function Tests | 1.8100000E-006 | - |
| ENSG00000149218 | Platelet Function Tests | 2.4400000E-007 | - |
| ENSG00000149218 | Platelet Function Tests | 2.7700000E-011 | - |
| ENSG00000149218 | Platelet Function Tests | 5.9900000E-011 | - |
| ENSG00000149218 | Platelet Function Tests | 1.0985900E-005 | - |
| ENSG00000149218 | Platelet Function Tests | 1.5054100E-005 | - |
| ENSG00000149218 | Platelet Function Tests | 3.9900000E-006 | - |
| ENSG00000149218 | Platelet Function Tests | 7.4900000E-007 | - |
| ENSG00000149218 | Platelet Function Tests | 2.8600000E-011 | - |
| ENSG00000149218 | Platelet Function Tests | 6.4350132E-006 | - |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000149218 | rs10605167 | 11 | 95095304 | T | Mean platelet volume | 27863252 | [0.031-0.049] unit decrease | 0.03997559 | EFO_0004584 |
| ENSG00000149218 | rs3740861 | 11 | 95129413 | G | Heel bone mineral density | 30598549 | [0.0078-0.0157] unit decrease | 0.0117704 | EFO_0009270 |
| ENSG00000149218 | rs3740861 | 11 | 95129413 | ? | Heel bone mineral density | 30595370 | EFO_0009270 |
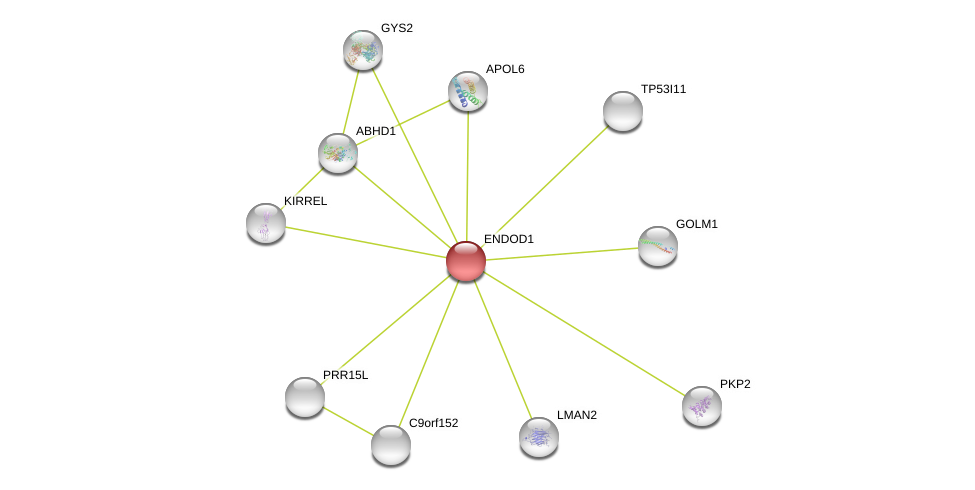
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000149218 | ENDOD1 | 80 | 74.314 | ENSAMEG00000019907 | ENDOD1 | 100 | 74.314 | Ailuropoda_melanoleuca |
| ENSG00000149218 | ENDOD1 | 51 | 30.847 | ENSAOCG00000021283 | si:dkey-85k7.10 | 95 | 30.847 | Amphiprion_ocellaris |
| ENSG00000149218 | ENDOD1 | 51 | 30.847 | ENSAPEG00000018856 | si:dkey-85k7.10 | 95 | 30.847 | Amphiprion_percula |
| ENSG00000149218 | ENDOD1 | 94 | 37.866 | ENSACAG00000013977 | ENDOD1 | 99 | 38.153 | Anolis_carolinensis |
| ENSG00000149218 | ENDOD1 | 52 | 31.034 | ENSAMXG00000037726 | si:dkey-85k7.10 | 88 | 31.970 | Astyanax_mexicanus |
| ENSG00000149218 | ENDOD1 | 100 | 93.800 | ENSCJAG00000039825 | ENDOD1 | 100 | 93.800 | Callithrix_jacchus |
| ENSG00000149218 | ENDOD1 | 100 | 92.600 | ENSCCAG00000036865 | ENDOD1 | 100 | 92.600 | Cebus_capucinus |
| ENSG00000149218 | ENDOD1 | 100 | 96.400 | ENSCATG00000044441 | ENDOD1 | 100 | 96.400 | Cercocebus_atys |
| ENSG00000149218 | ENDOD1 | 100 | 97.400 | ENSCSAG00000003290 | ENDOD1 | 100 | 97.400 | Chlorocebus_sabaeus |
| ENSG00000149218 | ENDOD1 | 68 | 83.529 | ENSCHOG00000013059 | ENDOD1 | 100 | 83.529 | Choloepus_hoffmanni |
| ENSG00000149218 | ENDOD1 | 55 | 31.475 | ENSCPBG00000005639 | - | 54 | 31.475 | Chrysemys_picta_bellii |
| ENSG00000149218 | ENDOD1 | 97 | 45.062 | ENSCPBG00000024041 | ENDOD1 | 98 | 44.856 | Chrysemys_picta_bellii |
| ENSG00000149218 | ENDOD1 | 61 | 31.818 | ENSCPBG00000009952 | - | 56 | 31.429 | Chrysemys_picta_bellii |
| ENSG00000149218 | ENDOD1 | 56 | 32.095 | ENSCPBG00000007740 | - | 65 | 32.095 | Chrysemys_picta_bellii |
| ENSG00000149218 | ENDOD1 | 100 | 97.200 | ENSCANG00000025695 | ENDOD1 | 100 | 97.200 | Colobus_angolensis_palliatus |
| ENSG00000149218 | ENDOD1 | 100 | 84.600 | ENSEASG00005011271 | ENDOD1 | 100 | 84.600 | Equus_asinus_asinus |
| ENSG00000149218 | ENDOD1 | 100 | 84.800 | ENSECAG00000017667 | ENDOD1 | 100 | 84.800 | Equus_caballus |
| ENSG00000149218 | ENDOD1 | 100 | 80.400 | ENSFCAG00000005079 | ENDOD1 | 100 | 80.400 | Felis_catus |
| ENSG00000149218 | ENDOD1 | 99 | 39.960 | ENSFALG00000000330 | ENDOD1 | 86 | 39.960 | Ficedula_albicollis |
| ENSG00000149218 | ENDOD1 | 80 | 84.040 | ENSFDAG00000012281 | ENDOD1 | 94 | 84.040 | Fukomys_damarensis |
| ENSG00000149218 | ENDOD1 | 93 | 41.453 | ENSGALG00000017205 | ENDOD1 | 93 | 41.453 | Gallus_gallus |
| ENSG00000149218 | ENDOD1 | 77 | 41.192 | ENSGAGG00000005154 | - | 99 | 40.933 | Gopherus_agassizii |
| ENSG00000149218 | ENDOD1 | 51 | 33.206 | ENSGAGG00000016583 | - | 66 | 33.206 | Gopherus_agassizii |
| ENSG00000149218 | ENDOD1 | 54 | 32.646 | ENSGAGG00000016563 | - | 80 | 33.740 | Gopherus_agassizii |
| ENSG00000149218 | ENDOD1 | 94 | 99.149 | ENSGGOG00000024775 | ENDOD1 | 99 | 97.468 | Gorilla_gorilla |
| ENSG00000149218 | ENDOD1 | 53 | 31.250 | ENSHBUG00000008382 | - | 97 | 31.250 | Haplochromis_burtoni |
| ENSG00000149218 | ENDOD1 | 100 | 83.600 | ENSHGLG00000010395 | ENDOD1 | 100 | 83.600 | Heterocephalus_glaber_female |
| ENSG00000149218 | ENDOD1 | 100 | 82.000 | ENSHGLG00100000983 | ENDOD1 | 100 | 82.000 | Heterocephalus_glaber_male |
| ENSG00000149218 | ENDOD1 | 100 | 84.600 | ENSSTOG00000000032 | ENDOD1 | 100 | 84.600 | Ictidomys_tridecemlineatus |
| ENSG00000149218 | ENDOD1 | 53 | 31.786 | ENSLACG00000005002 | - | 98 | 31.786 | Latimeria_chalumnae |
| ENSG00000149218 | ENDOD1 | 100 | 78.389 | ENSLAFG00000010482 | ENDOD1 | 100 | 78.389 | Loxodonta_africana |
| ENSG00000149218 | ENDOD1 | 100 | 96.600 | ENSMFAG00000037393 | ENDOD1 | 100 | 96.600 | Macaca_fascicularis |
| ENSG00000149218 | ENDOD1 | 100 | 97.000 | ENSMMUG00000047793 | ENDOD1 | 100 | 97.000 | Macaca_mulatta |
| ENSG00000149218 | ENDOD1 | 100 | 97.000 | ENSMNEG00000040186 | ENDOD1 | 100 | 97.000 | Macaca_nemestrina |
| ENSG00000149218 | ENDOD1 | 82 | 97.800 | ENSMLEG00000043710 | ENDOD1 | 100 | 97.800 | Mandrillus_leucophaeus |
| ENSG00000149218 | ENDOD1 | 100 | 66.733 | ENSMODG00000000165 | ENDOD1 | 100 | 66.733 | Monodelphis_domestica |
| ENSG00000149218 | ENDOD1 | 80 | 82.793 | ENSMLUG00000014495 | ENDOD1 | 100 | 82.793 | Myotis_lucifugus |
| ENSG00000149218 | ENDOD1 | 100 | 96.800 | ENSNLEG00000007127 | ENDOD1 | 100 | 96.800 | Nomascus_leucogenys |
| ENSG00000149218 | ENDOD1 | 100 | 67.331 | ENSMEUG00000008592 | ENDOD1 | 100 | 67.331 | Notamacropus_eugenii |
| ENSG00000149218 | ENDOD1 | 51 | 30.496 | ENSONIG00000014582 | - | 98 | 30.496 | Oreochromis_niloticus |
| ENSG00000149218 | ENDOD1 | 99 | 58.366 | ENSOANG00000010405 | - | 99 | 59.252 | Ornithorhynchus_anatinus |
| ENSG00000149218 | ENDOD1 | 99 | 60.800 | ENSOANG00000010505 | - | 100 | 60.800 | Ornithorhynchus_anatinus |
| ENSG00000149218 | ENDOD1 | 100 | 87.600 | ENSOCUG00000014040 | ENDOD1 | 100 | 87.600 | Oryctolagus_cuniculus |
| ENSG00000149218 | ENDOD1 | 53 | 30.717 | ENSOMEG00000007309 | - | 52 | 30.717 | Oryzias_melastigma |
| ENSG00000149218 | ENDOD1 | 96 | 86.694 | ENSOGAG00000026634 | ENDOD1 | 100 | 86.400 | Otolemur_garnettii |
| ENSG00000149218 | ENDOD1 | 100 | 99.200 | ENSPPAG00000036031 | ENDOD1 | 100 | 99.200 | Pan_paniscus |
| ENSG00000149218 | ENDOD1 | 100 | 80.800 | ENSPPRG00000004151 | - | 100 | 80.800 | Panthera_pardus |
| ENSG00000149218 | ENDOD1 | 84 | 76.850 | ENSPTIG00000021253 | ENDOD1 | 99 | 76.850 | Panthera_tigris_altaica |
| ENSG00000149218 | ENDOD1 | 100 | 99.400 | ENSPTRG00000045007 | ENDOD1 | 100 | 99.400 | Pan_troglodytes |
| ENSG00000149218 | ENDOD1 | 100 | 96.800 | ENSPANG00000009553 | ENDOD1 | 100 | 96.800 | Papio_anubis |
| ENSG00000149218 | ENDOD1 | 58 | 32.399 | ENSPKIG00000003602 | - | 54 | 32.288 | Paramormyrops_kingsleyae |
| ENSG00000149218 | ENDOD1 | 63 | 31.624 | ENSPKIG00000007324 | - | 50 | 32.770 | Paramormyrops_kingsleyae |
| ENSG00000149218 | ENDOD1 | 58 | 34.268 | ENSPKIG00000019032 | - | 54 | 34.169 | Paramormyrops_kingsleyae |
| ENSG00000149218 | ENDOD1 | 51 | 34.586 | ENSPSIG00000010161 | - | 91 | 34.733 | Pelodiscus_sinensis |
| ENSG00000149218 | ENDOD1 | 57 | 33.108 | ENSPSIG00000009892 | - | 99 | 33.108 | Pelodiscus_sinensis |
| ENSG00000149218 | ENDOD1 | 96 | 41.788 | ENSPSIG00000011246 | ENDOD1 | 100 | 41.788 | Pelodiscus_sinensis |
| ENSG00000149218 | ENDOD1 | 52 | 30.877 | ENSPMGG00000009447 | si:dkey-243k1.3 | 99 | 30.877 | Periophthalmus_magnuspinnatus |
| ENSG00000149218 | ENDOD1 | 100 | 68.924 | ENSPCIG00000001331 | ENDOD1 | 100 | 68.924 | Phascolarctos_cinereus |
| ENSG00000149218 | ENDOD1 | 53 | 31.673 | ENSPFOG00000007195 | - | 99 | 31.673 | Poecilia_formosa |
| ENSG00000149218 | ENDOD1 | 54 | 31.561 | ENSPREG00000018344 | - | 99 | 31.561 | Poecilia_reticulata |
| ENSG00000149218 | ENDOD1 | 100 | 83.800 | ENSPCAG00000000704 | ENDOD1 | 100 | 83.800 | Procavia_capensis |
| ENSG00000149218 | ENDOD1 | 100 | 87.200 | ENSPCOG00000008619 | ENDOD1 | 100 | 87.200 | Propithecus_coquereli |
| ENSG00000149218 | ENDOD1 | 56 | 30.721 | ENSPNYG00000003385 | si:ch211-165i18.2 | 86 | 32.411 | Pundamilia_nyererei |
| ENSG00000149218 | ENDOD1 | 53 | 30.137 | ENSPNAG00000025759 | - | 94 | 30.137 | Pygocentrus_nattereri |
| ENSG00000149218 | ENDOD1 | 53 | 31.186 | ENSPNAG00000004857 | si:dkey-85k7.10 | 95 | 31.186 | Pygocentrus_nattereri |
| ENSG00000149218 | ENDOD1 | 50 | 30.970 | ENSPNAG00000025288 | - | 94 | 30.970 | Pygocentrus_nattereri |
| ENSG00000149218 | ENDOD1 | 51 | 30.605 | ENSPNAG00000025281 | - | 93 | 30.605 | Pygocentrus_nattereri |
| ENSG00000149218 | ENDOD1 | 100 | 96.800 | ENSRBIG00000025792 | ENDOD1 | 100 | 96.800 | Rhinopithecus_bieti |
| ENSG00000149218 | ENDOD1 | 100 | 96.800 | ENSRROG00000025221 | ENDOD1 | 100 | 96.800 | Rhinopithecus_roxellana |
| ENSG00000149218 | ENDOD1 | 100 | 66.335 | ENSSHAG00000016362 | ENDOD1 | 100 | 66.335 | Sarcophilus_harrisii |
| ENSG00000149218 | ENDOD1 | 95 | 41.875 | ENSSPUG00000004451 | ENDOD1 | 97 | 42.062 | Sphenodon_punctatus |
| ENSG00000149218 | ENDOD1 | 52 | 30.208 | ENSTNIG00000004190 | si:dkey-85k7.10 | 97 | 30.208 | Tetraodon_nigroviridis |
| ENSG00000149218 | ENDOD1 | 90 | 87.054 | ENSTBEG00000014696 | ENDOD1 | 99 | 87.054 | Tupaia_belangeri |
| ENSG00000149218 | ENDOD1 | 69 | 73.121 | ENSUAMG00000007826 | - | 100 | 73.121 | Ursus_americanus |
| ENSG00000149218 | ENDOD1 | 69 | 73.121 | ENSUMAG00000007178 | ENDOD1 | 100 | 73.121 | Ursus_maritimus |
| ENSG00000149218 | ENDOD1 | 53 | 30.584 | ENSXETG00000030024 | - | 93 | 30.584 | Xenopus_tropicalis |
| ENSG00000149218 | ENDOD1 | 53 | 30.719 | ENSXMAG00000009699 | - | 94 | 31.293 | Xiphophorus_maculatus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0002576 | platelet degranulation | - | TAS | Process |
| GO:0003676 | nucleic acid binding | - | IEA | Function |
| GO:0004519 | endonuclease activity | - | IEA | Function |
| GO:0005576 | extracellular region | - | TAS | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0016020 | membrane | 19946888. | HDA | Component |
| GO:0046872 | metal ion binding | - | IEA | Function |
| GO:0070062 | extracellular exosome | 23533145. | HDA | Component |
| GO:0090305 | nucleic acid phosphodiester bond hydrolysis | - | IEA | Process |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| DLBC | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRP | |||||||
| KIRP | |||||||
| KIRP | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| ESCA | |||
| LUSC | |||
| TGCT | |||
| UCS |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| ACC | ||
| HNSC | ||
| BRCA | ||
| ESCA | ||
| PAAD | ||
| BLCA | ||
| CESC | ||
| GBM | ||
| LIHC | ||
| LGG | ||
| CHOL | ||
| THCA |