
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000536243 | PLCB3-203 | 471 | - | - | - (aa) | - | - |
| ENST00000540288 | PLCB3-204 | 4469 | XM_011545101 | ENSP00000443631 | 1234 (aa) | XP_011543403 | Q01970 |
| ENST00000325234 | PLCB3-202 | 3946 | - | ENSP00000324660 | 1167 (aa) | - | Q01970 |
| ENST00000279230 | PLCB3-201 | 4197 | - | ENSP00000279230 | 1234 (aa) | - | Q01970 |
| Pathway ID | Pathway Name | Source |
|---|---|---|
| hsa00562 | Inositol phosphate metabolism | KEGG |
| hsa01100 | Metabolic pathways | KEGG |
| hsa04015 | Rap1 signaling pathway | KEGG |
| hsa04020 | Calcium signaling pathway | KEGG |
| hsa04022 | cGMP-PKG signaling pathway | KEGG |
| hsa04062 | Chemokine signaling pathway | KEGG |
| hsa04070 | Phosphatidylinositol signaling system | KEGG |
| hsa04071 | Sphingolipid signaling pathway | KEGG |
| hsa04072 | Phospholipase D signaling pathway | KEGG |
| hsa04261 | Adrenergic signaling in cardiomyocytes | KEGG |
| hsa04270 | Vascular smooth muscle contraction | KEGG |
| hsa04310 | Wnt signaling pathway | KEGG |
| hsa04371 | Apelin signaling pathway | KEGG |
| hsa04540 | Gap junction | KEGG |
| hsa04611 | Platelet activation | KEGG |
| hsa04621 | NOD-like receptor signaling pathway | KEGG |
| hsa04713 | Circadian entrainment | KEGG |
| hsa04720 | Long-term potentiation | KEGG |
| hsa04723 | Retrograde endocannabinoid signaling | KEGG |
| hsa04724 | Glutamatergic synapse | KEGG |
| hsa04725 | Cholinergic synapse | KEGG |
| hsa04726 | Serotonergic synapse | KEGG |
| hsa04728 | Dopaminergic synapse | KEGG |
| hsa04730 | Long-term depression | KEGG |
| hsa04750 | Inflammatory mediator regulation of TRP channels | KEGG |
| hsa04911 | Insulin secretion | KEGG |
| hsa04912 | GnRH signaling pathway | KEGG |
| hsa04915 | Estrogen signaling pathway | KEGG |
| hsa04916 | Melanogenesis | KEGG |
| hsa04918 | Thyroid hormone synthesis | KEGG |
| hsa04919 | Thyroid hormone signaling pathway | KEGG |
| hsa04921 | Oxytocin signaling pathway | KEGG |
| hsa04922 | Glucagon signaling pathway | KEGG |
| hsa04924 | Renin secretion | KEGG |
| hsa04925 | Aldosterone synthesis and secretion | KEGG |
| hsa04926 | Relaxin signaling pathway | KEGG |
| hsa04927 | Cortisol synthesis and secretion | KEGG |
| hsa04928 | Parathyroid hormone synthesis, secretion and action | KEGG |
| hsa04933 | AGE-RAGE signaling pathway in diabetic complications | KEGG |
| hsa04934 | Cushing syndrome | KEGG |
| hsa04961 | Endocrine and other factor-regulated calcium reabsorption | KEGG |
| hsa04970 | Salivary secretion | KEGG |
| hsa04971 | Gastric acid secretion | KEGG |
| hsa04972 | Pancreatic secretion | KEGG |
| hsa05010 | Alzheimer disease | KEGG |
| hsa05016 | Huntington disease | KEGG |
| hsa05142 | Chagas disease (American trypanosomiasis) | KEGG |
| hsa05143 | African trypanosomiasis | KEGG |
| hsa05146 | Amoebiasis | KEGG |
| hsa05163 | Human cytomegalovirus infection | KEGG |
| hsa05200 | Pathways in cancer | KEGG |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000149782 | rs588177 | 11 | 64256584 | A | High light scatter reticulocyte percentage of red cells | 27863252 | [0.016-0.031] unit decrease | 0.0234532 | EFO_0007986 |
| ENSG00000149782 | rs12421615 | 11 | 64254133 | G | Vitiligo | 27723757 | [1.1-1.2] | 1.1498218 | EFO_0004208 |
| ENSG00000149782 | rs588177 | 11 | 64256584 | A | High light scatter reticulocyte count | 27863252 | [0.017-0.032] unit decrease | 0.0247408 | EFO_0007986 |
| ENSG00000149782 | rs588177 | 11 | 64256584 | ? | Systolic blood pressure | 30595370 | EFO_0006335 | ||
| ENSG00000149782 | rs117874826 | 11 | 64260194 | ? | Systolic blood pressure | 30595370 | EFO_0006335 | ||
| ENSG00000149782 | rs589030 | 11 | 64264326 | ? | Mean corpuscular hemoglobin | 30595370 | EFO_0004527 | ||
| ENSG00000149782 | rs589030 | 11 | 64264326 | ? | Red blood cell count | 30595370 | EFO_0004305 | ||
| ENSG00000149782 | rs35169799 | 11 | 64263769 | T | Waist-to-hip ratio adjusted for BMI (additive genetic model) | 30778226 | [0.025-0.043] unit increase | 0.0339 | EFO_0007788 |
| ENSG00000149782 | rs35169799 | 11 | 64263769 | T | Waist-to-hip ratio adjusted for BMI (additive genetic model) | 30778226 | [0.024-0.042] unit increase | 0.0327 | EFO_0007788 |
| ENSG00000149782 | rs35169799 | 11 | 64263769 | T | Waist-to-hip ratio adjusted for BMI (additive genetic model) | 30778226 | [0.037-0.061] unit increase | 0.0492 | EFO_0007788 |
| ENSG00000149782 | rs35169799 | 11 | 64263769 | T | Waist-to-hip ratio adjusted for BMI (additive genetic model) | 30778226 | [0.037-0.061] unit increase | 0.0489 | EFO_0007788 |
| ENSG00000149782 | rs35169799 | 11 | 64263769 | T | Type 2 diabetes (adjusted for BMI) | 29632382 | [1.03-1.08] | 1.05 | EFO_0001360 |
| ENSG00000149782 | rs35169799 | 11 | 64263769 | T | Type 2 diabetes (adjusted for BMI) | 29632382 | [1.03-1.09] | 1.06 | EFO_0001360 |
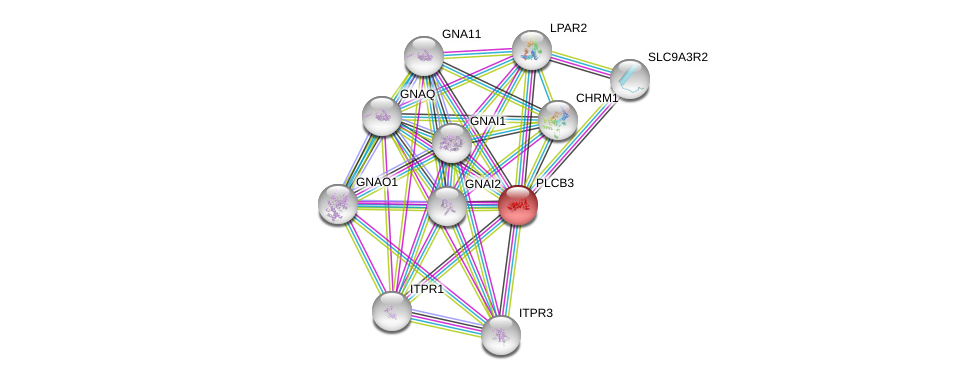
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000149782 | PLCB3 | 100 | 92.227 | ENSMUSG00000024960 | Plcb3 | 100 | 92.227 | Mus_musculus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0003073 | regulation of systemic arterial blood pressure | 18468998. | IDA | Process |
| GO:0004435 | phosphatidylinositol phospholipase C activity | 21873635. | IBA | Function |
| GO:0004629 | phospholipase C activity | - | TAS | Function |
| GO:0005509 | calcium ion binding | - | IEA | Function |
| GO:0005515 | protein binding | 21873635. | IBA | Function |
| GO:0005515 | protein binding | 21822282.23377541. | IPI | Function |
| GO:0005516 | calmodulin binding | 21873635. | IBA | Function |
| GO:0005516 | calmodulin binding | 12821674. | IDA | Function |
| GO:0005654 | nucleoplasm | - | IDA | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0007186 | G protein-coupled receptor signaling pathway | - | TAS | Process |
| GO:0007223 | Wnt signaling pathway, calcium modulating pathway | - | TAS | Process |
| GO:0016020 | membrane | - | IEA | Component |
| GO:0016042 | lipid catabolic process | - | IEA | Process |
| GO:0032959 | inositol trisphosphate biosynthetic process | 21873635. | IBA | Process |
| GO:0032991 | protein-containing complex | 18468998. | IDA | Component |
| GO:0043647 | inositol phosphate metabolic process | - | TAS | Process |
| GO:0045296 | cadherin binding | 25468996. | HDA | Function |
| GO:0048015 | phosphatidylinositol-mediated signaling | 21873635. | IBA | Process |
| GO:0051209 | release of sequestered calcium ion into cytosol | 21873635. | IBA | Process |
| GO:0099524 | postsynaptic cytosol | - | IEA | Component |