
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000281589 | PABP | PF00658.18 | 3.8e-30 | 1 | 1 |
| ENSP00000281589 | RRM_5 | PF13893.6 | 5.2e-08 | 1 | 2 |
| ENSP00000281589 | RRM_5 | PF13893.6 | 5.2e-08 | 2 | 2 |
| ENSP00000281589 | RRM_1 | PF00076.22 | 5.3e-89 | 1 | 4 |
| ENSP00000281589 | RRM_1 | PF00076.22 | 5.3e-89 | 2 | 4 |
| ENSP00000281589 | RRM_1 | PF00076.22 | 5.3e-89 | 3 | 4 |
| ENSP00000281589 | RRM_1 | PF00076.22 | 5.3e-89 | 4 | 4 |
| PID | Title | Method | Time | Author | Doi |
|---|---|---|---|---|---|
| 29431736 |
| PID | Title | Article | Time | Author | Doi |
|---|---|---|---|---|---|
| 27802063 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000281589 | PABPC3-201 | 3387 | - | ENSP00000281589 | 631 (aa) | - | Q9H361 |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000151846 | Body Mass Index | 3.69127265951175E-5 | 17903300 |
| ENSG00000151846 | Body Mass Index | 9.1221293583299E-6 | 17903300 |
| ENSG00000151846 | Body Weight Changes | 9.94212450016185E-5 | 17903300 |
| ENSG00000151846 | Body Mass Index | 5.62533436854196E-5 | 17903300 |
| ENSG00000151846 | Body Mass Index | 6.70008838343783E-6 | 17903300 |
| ENSG00000151846 | Body Weight Changes | 3.72348892492269E-5 | 17903300 |
| ENSG00000151846 | Body Weight | 4.803246198537E-5 | 17903300 |
| ENSG00000151846 | Body Weight | 3.33930811104999E-5 | 17903300 |
| ENSG00000151846 | Glomerular Filtration Rate | 1.43242652486927E-5 | 17903292 |
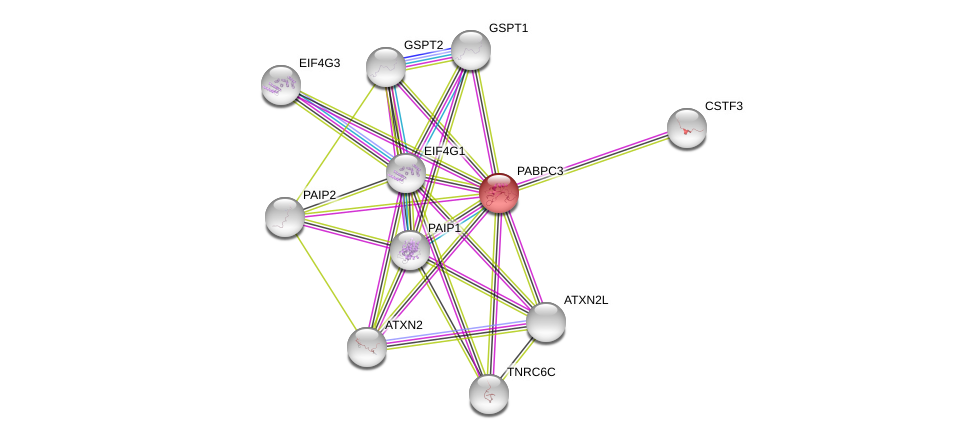
| Ensembl ID | Gene Symbol | Coverage | Identiy | Paralog | Gene Symbol | Coverage | Identiy |
|---|---|---|---|---|---|---|---|
| ENSG00000151846 | PABPC3 | 54 | 30.189 | ENSG00000147140 | NONO | 73 | 30.189 |
| ENSG00000151846 | PABPC3 | 55 | 30.588 | ENSG00000106344 | RBM28 | 64 | 31.061 |
| ENSG00000151846 | PABPC3 | 98 | 65.862 | ENSG00000101104 | PABPC1L | 98 | 73.765 |
| ENSG00000151846 | PABPC3 | 58 | 62.360 | ENSG00000174740 | PABPC5 | 100 | 59.155 |
| ENSG00000151846 | PABPC3 | 51 | 42.857 | ENSG00000099783 | HNRNPM | 61 | 30.000 |
| ENSG00000151846 | PABPC3 | 58 | 32.099 | ENSG00000066044 | ELAVL1 | 82 | 32.099 |
| ENSG00000151846 | PABPC3 | 61 | 32.450 | ENSG00000107105 | ELAVL2 | 89 | 32.450 |
| ENSG00000151846 | PABPC3 | 56 | 37.500 | ENSG00000100461 | RBM23 | 66 | 37.500 |
| ENSG00000151846 | PABPC3 | 66 | 30.204 | ENSG00000116001 | TIA1 | 88 | 37.226 |
| ENSG00000151846 | PABPC3 | 59 | 34.722 | ENSG00000196361 | ELAVL3 | 73 | 34.722 |
| ENSG00000151846 | PABPC3 | 58 | 73.333 | ENSG00000186288 | PABPC1L2A | 98 | 73.333 |
| ENSG00000151846 | PABPC3 | 53 | 30.435 | ENSG00000179950 | PUF60 | 62 | 30.435 |
| ENSG00000151846 | PABPC3 | 58 | 36.612 | ENSG00000143368 | SF3B4 | 77 | 36.000 |
| ENSG00000151846 | PABPC3 | 61 | 67.838 | ENSG00000254535 | PABPC4L | 99 | 67.838 |
| ENSG00000151846 | PABPC3 | 58 | 32.203 | ENSG00000180098 | TRNAU1AP | 71 | 32.203 |
| ENSG00000151846 | PABPC3 | 99 | 72.671 | ENSG00000090621 | PABPC4 | 99 | 85.586 |
| ENSG00000151846 | PABPC3 | 63 | 31.579 | ENSG00000162374 | ELAVL4 | 88 | 31.579 |
| ENSG00000151846 | PABPC3 | 56 | 37.692 | ENSG00000151923 | TIAL1 | 98 | 37.692 |
| ENSG00000151846 | PABPC3 | 57 | 37.500 | ENSG00000131051 | RBM39 | 79 | 37.500 |
| ENSG00000151846 | PABPC3 | 100 | 91.994 | ENSG00000070756 | PABPC1 | 98 | 97.561 |
| ENSG00000151846 | PABPC3 | 58 | 73.333 | ENSG00000184388 | PABPC1L2B | 98 | 73.333 |
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000151846 | PABPC3 | 100 | 91.523 | ENSAMEG00000000348 | PABPC1 | 100 | 91.523 | Ailuropoda_melanoleuca |
| ENSG00000151846 | PABPC3 | 99 | 90.767 | ENSAPLG00000001129 | PABPC1 | 100 | 90.941 | Anas_platyrhynchos |
| ENSG00000151846 | PABPC3 | 100 | 85.460 | ENSACAG00000016470 | PABPC1 | 100 | 85.460 | Anolis_carolinensis |
| ENSG00000151846 | PABPC3 | 100 | 91.837 | ENSANAG00000033352 | - | 100 | 91.837 | Aotus_nancymaae |
| ENSG00000151846 | PABPC3 | 100 | 87.639 | ENSANAG00000027730 | - | 100 | 87.639 | Aotus_nancymaae |
| ENSG00000151846 | PABPC3 | 100 | 75.829 | ENSANAG00000026678 | - | 100 | 75.987 | Aotus_nancymaae |
| ENSG00000151846 | PABPC3 | 99 | 71.815 | ENSANAG00000035841 | - | 99 | 72.459 | Aotus_nancymaae |
| ENSG00000151846 | PABPC3 | 88 | 77.617 | ENSANAG00000019222 | - | 100 | 77.617 | Aotus_nancymaae |
| ENSG00000151846 | PABPC3 | 99 | 70.826 | ENSANAG00000036324 | - | 99 | 69.913 | Aotus_nancymaae |
| ENSG00000151846 | PABPC3 | 100 | 83.542 | ENSAMXG00000018395 | - | 100 | 83.542 | Astyanax_mexicanus |
| ENSG00000151846 | PABPC3 | 100 | 91.994 | ENSBTAG00000046358 | PABPC1 | 100 | 91.994 | Bos_taurus |
| ENSG00000151846 | PABPC3 | 88 | 77.818 | ENSCJAG00000045282 | - | 99 | 77.818 | Callithrix_jacchus |
| ENSG00000151846 | PABPC3 | 71 | 89.868 | ENSCJAG00000041162 | - | 91 | 89.868 | Callithrix_jacchus |
| ENSG00000151846 | PABPC3 | 100 | 91.837 | ENSCJAG00000015833 | PABPC1 | 100 | 91.837 | Callithrix_jacchus |
| ENSG00000151846 | PABPC3 | 100 | 91.994 | ENSCAFG00000000567 | PABPC1 | 100 | 91.994 | Canis_familiaris |
| ENSG00000151846 | PABPC3 | 100 | 91.994 | ENSCAFG00020000440 | PABPC1 | 100 | 91.994 | Canis_lupus_dingo |
| ENSG00000151846 | PABPC3 | 100 | 92.710 | ENSCHIG00000023714 | PABPC1 | 100 | 92.710 | Capra_hircus |
| ENSG00000151846 | PABPC3 | 100 | 79.528 | ENSTSYG00000037130 | - | 100 | 79.528 | Carlito_syrichta |
| ENSG00000151846 | PABPC3 | 70 | 82.432 | ENSTSYG00000033737 | - | 90 | 82.658 | Carlito_syrichta |
| ENSG00000151846 | PABPC3 | 100 | 91.994 | ENSTSYG00000004229 | PABPC1 | 100 | 91.994 | Carlito_syrichta |
| ENSG00000151846 | PABPC3 | 95 | 91.367 | ENSCAPG00000013098 | PABPC1 | 98 | 91.367 | Cavia_aperea |
| ENSG00000151846 | PABPC3 | 99 | 93.019 | ENSCPOG00000012917 | PABPC1 | 98 | 93.019 | Cavia_porcellus |
| ENSG00000151846 | PABPC3 | 100 | 72.571 | ENSCCAG00000027395 | - | 100 | 72.571 | Cebus_capucinus |
| ENSG00000151846 | PABPC3 | 83 | 76.842 | ENSCCAG00000028512 | - | 98 | 74.429 | Cebus_capucinus |
| ENSG00000151846 | PABPC3 | 88 | 80.583 | ENSCCAG00000031444 | - | 100 | 80.583 | Cebus_capucinus |
| ENSG00000151846 | PABPC3 | 92 | 80.172 | ENSCCAG00000031750 | - | 97 | 80.172 | Cebus_capucinus |
| ENSG00000151846 | PABPC3 | 99 | 87.362 | ENSCCAG00000037208 | PABPC3 | 100 | 87.362 | Cebus_capucinus |
| ENSG00000151846 | PABPC3 | 95 | 90.952 | ENSCCAG00000037733 | - | 100 | 90.952 | Cebus_capucinus |
| ENSG00000151846 | PABPC3 | 100 | 93.344 | ENSCATG00000029313 | PABPC3 | 100 | 93.344 | Cercocebus_atys |
| ENSG00000151846 | PABPC3 | 99 | 71.672 | ENSCATG00000045341 | - | 99 | 76.068 | Cercocebus_atys |
| ENSG00000151846 | PABPC3 | 100 | 91.994 | ENSCATG00000039184 | PABPC1 | 100 | 91.994 | Cercocebus_atys |
| ENSG00000151846 | PABPC3 | 100 | 72.108 | ENSCATG00000044984 | - | 100 | 82.474 | Cercocebus_atys |
| ENSG00000151846 | PABPC3 | 99 | 92.832 | ENSCLAG00000009676 | PABPC1 | 100 | 92.832 | Chinchilla_lanigera |
| ENSG00000151846 | PABPC3 | 100 | 91.994 | ENSCSAG00000012017 | PABPC1 | 100 | 91.994 | Chlorocebus_sabaeus |
| ENSG00000151846 | PABPC3 | 99 | 80.660 | ENSCHOG00000008178 | PABPC1 | 99 | 80.660 | Choloepus_hoffmanni |
| ENSG00000151846 | PABPC3 | 100 | 89.953 | ENSCPBG00000005782 | PABPC1 | 100 | 89.953 | Chrysemys_picta_bellii |
| ENSG00000151846 | PABPC3 | 95 | 92.388 | ENSCANG00000021524 | PABPC1 | 97 | 92.388 | Colobus_angolensis_palliatus |
| ENSG00000151846 | PABPC3 | 100 | 91.601 | ENSCANG00000041712 | PABPC3 | 100 | 91.601 | Colobus_angolensis_palliatus |
| ENSG00000151846 | PABPC3 | 61 | 84.646 | ENSCANG00000036233 | - | 100 | 84.646 | Colobus_angolensis_palliatus |
| ENSG00000151846 | PABPC3 | 99 | 75.926 | ENSCGRG00001007983 | Pabpc6 | 99 | 75.772 | Cricetulus_griseus_chok1gshd |
| ENSG00000151846 | PABPC3 | 100 | 91.680 | ENSCGRG00001022067 | - | 100 | 91.680 | Cricetulus_griseus_chok1gshd |
| ENSG00000151846 | PABPC3 | 98 | 78.558 | ENSCGRG00000005826 | - | 89 | 78.752 | Cricetulus_griseus_crigri |
| ENSG00000151846 | PABPC3 | 99 | 75.926 | ENSCGRG00000019560 | Pabpc6 | 99 | 75.772 | Cricetulus_griseus_crigri |
| ENSG00000151846 | PABPC3 | 99 | 92.133 | ENSCGRG00000006487 | - | 100 | 92.133 | Cricetulus_griseus_crigri |
| ENSG00000151846 | PABPC3 | 100 | 91.994 | ENSDNOG00000008718 | PABPC1 | 100 | 91.994 | Dasypus_novemcinctus |
| ENSG00000151846 | PABPC3 | 97 | 65.116 | ENSDNOG00000030956 | - | 99 | 65.282 | Dasypus_novemcinctus |
| ENSG00000151846 | PABPC3 | 72 | 94.000 | ENSDNOG00000006598 | - | 100 | 94.000 | Dasypus_novemcinctus |
| ENSG00000151846 | PABPC3 | 100 | 91.837 | ENSDORG00000013935 | Pabpc1 | 100 | 91.837 | Dipodomys_ordii |
| ENSG00000151846 | PABPC3 | 96 | 84.494 | ENSETEG00000019213 | - | 100 | 84.719 | Echinops_telfairi |
| ENSG00000151846 | PABPC3 | 96 | 94.595 | ENSETEG00000002793 | - | 84 | 94.595 | Echinops_telfairi |
| ENSG00000151846 | PABPC3 | 100 | 91.994 | ENSEASG00005002810 | PABPC1 | 100 | 91.994 | Equus_asinus_asinus |
| ENSG00000151846 | PABPC3 | 98 | 47.896 | ENSEASG00005014126 | - | 97 | 47.727 | Equus_asinus_asinus |
| ENSG00000151846 | PABPC3 | 99 | 47.434 | ENSECAG00000038200 | - | 97 | 47.270 | Equus_caballus |
| ENSG00000151846 | PABPC3 | 100 | 91.994 | ENSECAG00000020699 | PABPC1 | 100 | 91.994 | Equus_caballus |
| ENSG00000151846 | PABPC3 | 99 | 91.930 | ENSEEUG00000002701 | PABPC1 | 99 | 91.930 | Erinaceus_europaeus |
| ENSG00000151846 | PABPC3 | 58 | 57.097 | ENSELUG00000005247 | - | 83 | 57.097 | Esox_lucius |
| ENSG00000151846 | PABPC3 | 100 | 91.994 | ENSFCAG00000001874 | PABPC1 | 100 | 92.698 | Felis_catus |
| ENSG00000151846 | PABPC3 | 99 | 90.767 | ENSFALG00000005841 | PABPC1 | 100 | 90.941 | Ficedula_albicollis |
| ENSG00000151846 | PABPC3 | 100 | 82.261 | ENSFDAG00000018480 | PABPC3 | 100 | 82.261 | Fukomys_damarensis |
| ENSG00000151846 | PABPC3 | 99 | 93.007 | ENSFDAG00000005583 | PABPC1 | 100 | 93.007 | Fukomys_damarensis |
| ENSG00000151846 | PABPC3 | 100 | 89.969 | ENSGALG00000030280 | PABPC1 | 100 | 90.125 | Gallus_gallus |
| ENSG00000151846 | PABPC3 | 100 | 90.581 | ENSGAGG00000000869 | PABPC1 | 100 | 90.581 | Gopherus_agassizii |
| ENSG00000151846 | PABPC3 | 99 | 93.007 | ENSGGOG00000006785 | - | 100 | 93.007 | Gorilla_gorilla |
| ENSG00000151846 | PABPC3 | 100 | 96.830 | ENSGGOG00000013146 | PABPC3 | 100 | 96.830 | Gorilla_gorilla |
| ENSG00000151846 | PABPC3 | 100 | 64.715 | ENSGGOG00000039851 | - | 100 | 65.077 | Gorilla_gorilla |
| ENSG00000151846 | PABPC3 | 100 | 59.862 | ENSGGOG00000041637 | - | 100 | 67.670 | Gorilla_gorilla |
| ENSG00000151846 | PABPC3 | 98 | 73.252 | ENSHGLG00000003042 | - | 97 | 73.252 | Heterocephalus_glaber_female |
| ENSG00000151846 | PABPC3 | 100 | 91.837 | ENSHGLG00000007713 | - | 100 | 91.837 | Heterocephalus_glaber_female |
| ENSG00000151846 | PABPC3 | 99 | 82.051 | ENSHGLG00100000417 | - | 99 | 81.513 | Heterocephalus_glaber_male |
| ENSG00000151846 | PABPC3 | 99 | 92.832 | ENSHGLG00100007448 | - | 100 | 92.832 | Heterocephalus_glaber_male |
| ENSG00000151846 | PABPC3 | 68 | 47.569 | ENSIPUG00000024284 | - | 95 | 47.569 | Ictalurus_punctatus |
| ENSG00000151846 | PABPC3 | 53 | 59.740 | ENSIPUG00000020086 | - | 85 | 59.740 | Ictalurus_punctatus |
| ENSG00000151846 | PABPC3 | 65 | 50.000 | ENSIPUG00000019895 | - | 60 | 50.000 | Ictalurus_punctatus |
| ENSG00000151846 | PABPC3 | 95 | 91.763 | ENSSTOG00000016395 | PABPC3 | 100 | 91.763 | Ictidomys_tridecemlineatus |
| ENSG00000151846 | PABPC3 | 100 | 74.254 | ENSJJAG00000014258 | Pabpc2 | 100 | 74.254 | Jaculus_jaculus |
| ENSG00000151846 | PABPC3 | 100 | 91.994 | ENSJJAG00000015918 | Pabpc1 | 100 | 91.994 | Jaculus_jaculus |
| ENSG00000151846 | PABPC3 | 99 | 74.519 | ENSJJAG00000019230 | Pabpc6 | 100 | 74.519 | Jaculus_jaculus |
| ENSG00000151846 | PABPC3 | 100 | 89.498 | ENSLACG00000004395 | PABPC1 | 100 | 89.498 | Latimeria_chalumnae |
| ENSG00000151846 | PABPC3 | 100 | 85.938 | ENSLOCG00000009681 | PABPC1 | 100 | 86.094 | Lepisosteus_oculatus |
| ENSG00000151846 | PABPC3 | 100 | 64.050 | ENSLAFG00000007939 | - | 100 | 64.364 | Loxodonta_africana |
| ENSG00000151846 | PABPC3 | 100 | 91.994 | ENSLAFG00000013994 | PABPC1 | 100 | 91.994 | Loxodonta_africana |
| ENSG00000151846 | PABPC3 | 99 | 68.654 | ENSMFAG00000034213 | - | 100 | 69.506 | Macaca_fascicularis |
| ENSG00000151846 | PABPC3 | 100 | 91.994 | ENSMFAG00000044845 | PABPC1 | 100 | 91.994 | Macaca_fascicularis |
| ENSG00000151846 | PABPC3 | 100 | 93.819 | ENSMFAG00000016858 | PABPC3 | 100 | 93.819 | Macaca_fascicularis |
| ENSG00000151846 | PABPC3 | 99 | 92.334 | ENSMFAG00000004725 | - | 99 | 92.334 | Macaca_fascicularis |
| ENSG00000151846 | PABPC3 | 100 | 90.293 | ENSMMUG00000017786 | PABPC1 | 100 | 90.952 | Macaca_mulatta |
| ENSG00000151846 | PABPC3 | 99 | 64.976 | ENSMMUG00000041053 | - | 100 | 65.610 | Macaca_mulatta |
| ENSG00000151846 | PABPC3 | 99 | 92.160 | ENSMMUG00000047411 | - | 98 | 92.174 | Macaca_mulatta |
| ENSG00000151846 | PABPC3 | 69 | 92.971 | ENSMMUG00000020835 | PABPC3 | 93 | 92.971 | Macaca_mulatta |
| ENSG00000151846 | PABPC3 | 99 | 92.160 | ENSMNEG00000010564 | - | 99 | 92.160 | Macaca_nemestrina |
| ENSG00000151846 | PABPC3 | 99 | 83.438 | ENSMNEG00000009340 | - | 100 | 91.078 | Macaca_nemestrina |
| ENSG00000151846 | PABPC3 | 100 | 66.719 | ENSMNEG00000007852 | - | 100 | 66.719 | Macaca_nemestrina |
| ENSG00000151846 | PABPC3 | 100 | 94.295 | ENSMNEG00000042936 | PABPC3 | 100 | 94.295 | Macaca_nemestrina |
| ENSG00000151846 | PABPC3 | 100 | 93.502 | ENSMLEG00000042214 | PABPC3 | 100 | 93.978 | Mandrillus_leucophaeus |
| ENSG00000151846 | PABPC3 | 99 | 92.845 | ENSMLEG00000027792 | - | 97 | 92.845 | Mandrillus_leucophaeus |
| ENSG00000151846 | PABPC3 | 85 | 74.535 | ENSMLEG00000043225 | - | 83 | 74.164 | Mandrillus_leucophaeus |
| ENSG00000151846 | PABPC3 | 95 | 65.565 | ENSMLEG00000041649 | - | 99 | 82.222 | Mandrillus_leucophaeus |
| ENSG00000151846 | PABPC3 | 99 | 90.000 | ENSMGAG00000012239 | PABPC1 | 100 | 90.172 | Meleagris_gallopavo |
| ENSG00000151846 | PABPC3 | 99 | 75.078 | ENSMAUG00000003031 | Pabpc6 | 100 | 75.117 | Mesocricetus_auratus |
| ENSG00000151846 | PABPC3 | 100 | 87.774 | ENSMAUG00000003523 | Pabpc1 | 100 | 87.774 | Mesocricetus_auratus |
| ENSG00000151846 | PABPC3 | 100 | 76.609 | ENSMAUG00000017991 | Pabpc2 | 100 | 77.237 | Mesocricetus_auratus |
| ENSG00000151846 | PABPC3 | 100 | 86.371 | ENSMICG00000011830 | PABPC3 | 100 | 86.529 | Microcebus_murinus |
| ENSG00000151846 | PABPC3 | 100 | 91.994 | ENSMICG00000009121 | PABPC1 | 100 | 91.994 | Microcebus_murinus |
| ENSG00000151846 | PABPC3 | 83 | 95.918 | ENSMOCG00000018191 | - | 100 | 95.918 | Microtus_ochrogaster |
| ENSG00000151846 | PABPC3 | 100 | 90.738 | ENSMOCG00000004921 | - | 100 | 90.738 | Microtus_ochrogaster |
| ENSG00000151846 | PABPC3 | 99 | 76.032 | ENSMOCG00000001354 | Pabpc2 | 99 | 76.190 | Microtus_ochrogaster |
| ENSG00000151846 | PABPC3 | 100 | 91.352 | ENSMODG00000006222 | PABPC1 | 100 | 91.352 | Monodelphis_domestica |
| ENSG00000151846 | PABPC3 | 100 | 83.465 | ENSMODG00000017105 | - | 100 | 83.465 | Monodelphis_domestica |
| ENSG00000151846 | PABPC3 | 99 | 84.348 | ENSMODG00000012040 | - | 99 | 84.348 | Monodelphis_domestica |
| ENSG00000151846 | PABPC3 | 99 | 76.161 | MGP_CAROLIEiJ_G0021058 | Pabpc6 | 99 | 76.006 | Mus_caroli |
| ENSG00000151846 | PABPC3 | 100 | 74.686 | MGP_CAROLIEiJ_G0022217 | Pabpc2 | 100 | 74.686 | Mus_caroli |
| ENSG00000151846 | PABPC3 | 100 | 92.284 | MGP_CAROLIEiJ_G0019740 | Pabpc1 | 100 | 93.162 | Mus_caroli |
| ENSG00000151846 | PABPC3 | 99 | 75.851 | ENSMUSG00000046173 | Pabpc6 | 99 | 75.851 | Mus_musculus |
| ENSG00000151846 | PABPC3 | 100 | 74.803 | ENSMUSG00000051732 | Pabpc2 | 100 | 74.803 | Mus_musculus |
| ENSG00000151846 | PABPC3 | 100 | 91.680 | ENSMUSG00000022283 | Pabpc1 | 99 | 93.162 | Mus_musculus |
| ENSG00000151846 | PABPC3 | 96 | 74.388 | MGP_PahariEiJ_G0018950 | Pabpc2 | 96 | 74.388 | Mus_pahari |
| ENSG00000151846 | PABPC3 | 85 | 68.698 | MGP_PahariEiJ_G0023374 | - | 99 | 68.975 | Mus_pahari |
| ENSG00000151846 | PABPC3 | 100 | 91.209 | MGP_PahariEiJ_G0019749 | Pabpc1 | 100 | 92.308 | Mus_pahari |
| ENSG00000151846 | PABPC3 | 100 | 91.680 | MGP_SPRETEiJ_G0020636 | Pabpc1 | 100 | 93.162 | Mus_spretus |
| ENSG00000151846 | PABPC3 | 100 | 74.371 | MGP_SPRETEiJ_G0023126 | Pabpc2 | 100 | 74.371 | Mus_spretus |
| ENSG00000151846 | PABPC3 | 99 | 75.542 | MGP_SPRETEiJ_G0021962 | Pabpc6 | 99 | 75.697 | Mus_spretus |
| ENSG00000151846 | PABPC3 | 99 | 92.670 | ENSMPUG00000015163 | PABPC1 | 100 | 92.670 | Mustela_putorius_furo |
| ENSG00000151846 | PABPC3 | 99 | 57.009 | ENSMLUG00000030069 | - | 99 | 57.232 | Myotis_lucifugus |
| ENSG00000151846 | PABPC3 | 99 | 88.088 | ENSMLUG00000012245 | PABPC1 | 100 | 88.088 | Myotis_lucifugus |
| ENSG00000151846 | PABPC3 | 99 | 74.643 | ENSNGAG00000000987 | Pabpc6 | 99 | 74.643 | Nannospalax_galili |
| ENSG00000151846 | PABPC3 | 99 | 69.949 | ENSNGAG00000023773 | - | 100 | 70.798 | Nannospalax_galili |
| ENSG00000151846 | PABPC3 | 100 | 91.994 | ENSNGAG00000000996 | Pabpc1 | 100 | 91.994 | Nannospalax_galili |
| ENSG00000151846 | PABPC3 | 99 | 80.604 | ENSNGAG00000023820 | - | 100 | 80.604 | Nannospalax_galili |
| ENSG00000151846 | PABPC3 | 100 | 74.254 | ENSNGAG00000016040 | Pabpc2 | 100 | 74.411 | Nannospalax_galili |
| ENSG00000151846 | PABPC3 | 68 | 62.284 | ENSNLEG00000034154 | - | 99 | 62.284 | Nomascus_leucogenys |
| ENSG00000151846 | PABPC3 | 100 | 67.512 | ENSNLEG00000030011 | - | 99 | 77.778 | Nomascus_leucogenys |
| ENSG00000151846 | PABPC3 | 98 | 79.008 | ENSNLEG00000034666 | - | 95 | 78.286 | Nomascus_leucogenys |
| ENSG00000151846 | PABPC3 | 100 | 92.393 | ENSNLEG00000018338 | PABPC3 | 99 | 92.393 | Nomascus_leucogenys |
| ENSG00000151846 | PABPC3 | 100 | 91.994 | ENSNLEG00000003142 | - | 100 | 91.994 | Nomascus_leucogenys |
| ENSG00000151846 | PABPC3 | 100 | 91.837 | ENSOPRG00000009667 | PABPC3 | 100 | 91.837 | Ochotona_princeps |
| ENSG00000151846 | PABPC3 | 99 | 91.400 | ENSODEG00000003229 | PABPC1 | 95 | 91.400 | Octodon_degus |
| ENSG00000151846 | PABPC3 | 77 | 83.333 | ENSODEG00000013269 | - | 96 | 83.333 | Octodon_degus |
| ENSG00000151846 | PABPC3 | 100 | 91.366 | ENSOANG00000007307 | PABPC1 | 100 | 91.366 | Ornithorhynchus_anatinus |
| ENSG00000151846 | PABPC3 | 100 | 91.837 | ENSOCUG00000004354 | - | 100 | 91.837 | Oryctolagus_cuniculus |
| ENSG00000151846 | PABPC3 | 100 | 91.680 | ENSOCUG00000023935 | - | 100 | 91.680 | Oryctolagus_cuniculus |
| ENSG00000151846 | PABPC3 | 99 | 75.517 | ENSOGAG00000033369 | - | 100 | 75.517 | Otolemur_garnettii |
| ENSG00000151846 | PABPC3 | 93 | 80.303 | ENSOGAG00000007921 | - | 100 | 80.303 | Otolemur_garnettii |
| ENSG00000151846 | PABPC3 | 100 | 84.977 | ENSOGAG00000033115 | - | 100 | 85.290 | Otolemur_garnettii |
| ENSG00000151846 | PABPC3 | 100 | 80.249 | ENSOGAG00000025522 | - | 100 | 80.249 | Otolemur_garnettii |
| ENSG00000151846 | PABPC3 | 99 | 93.019 | ENSOARG00000000095 | PABPC1 | 100 | 93.019 | Ovis_aries |
| ENSG00000151846 | PABPC3 | 99 | 61.982 | ENSPPAG00000036729 | - | 99 | 69.572 | Pan_paniscus |
| ENSG00000151846 | PABPC3 | 99 | 90.686 | ENSPPAG00000037498 | - | 100 | 90.826 | Pan_paniscus |
| ENSG00000151846 | PABPC3 | 100 | 66.301 | ENSPPAG00000037955 | - | 100 | 77.632 | Pan_paniscus |
| ENSG00000151846 | PABPC3 | 100 | 91.994 | ENSPPRG00000003755 | PABPC1 | 100 | 92.800 | Panthera_pardus |
| ENSG00000151846 | PABPC3 | 99 | 93.019 | ENSPTIG00000021083 | PABPC1 | 96 | 93.019 | Panthera_tigris_altaica |
| ENSG00000151846 | PABPC3 | 100 | 97.472 | ENSPTRG00000022593 | PABPC3 | 100 | 97.472 | Pan_troglodytes |
| ENSG00000151846 | PABPC3 | 100 | 91.994 | ENSPTRG00000047939 | PABPC1 | 100 | 92.810 | Pan_troglodytes |
| ENSG00000151846 | PABPC3 | 58 | 78.289 | ENSPTRG00000045319 | - | 100 | 77.961 | Pan_troglodytes |
| ENSG00000151846 | PABPC3 | 58 | 79.739 | ENSPTRG00000047501 | - | 100 | 80.392 | Pan_troglodytes |
| ENSG00000151846 | PABPC3 | 100 | 90.293 | ENSPANG00000010645 | PABPC1 | 100 | 90.293 | Papio_anubis |
| ENSG00000151846 | PABPC3 | 100 | 93.185 | ENSPANG00000032213 | PABPC3 | 100 | 93.185 | Papio_anubis |
| ENSG00000151846 | PABPC3 | 95 | 66.667 | ENSPANG00000030537 | - | 100 | 84.444 | Papio_anubis |
| ENSG00000151846 | PABPC3 | 72 | 91.139 | ENSPANG00000032575 | - | 100 | 91.139 | Papio_anubis |
| ENSG00000151846 | PABPC3 | 62 | 80.258 | ENSPKIG00000016835 | - | 97 | 80.258 | Paramormyrops_kingsleyae |
| ENSG00000151846 | PABPC3 | 59 | 75.000 | ENSPKIG00000002089 | - | 94 | 75.000 | Paramormyrops_kingsleyae |
| ENSG00000151846 | PABPC3 | 59 | 75.000 | ENSPKIG00000002905 | - | 94 | 75.000 | Paramormyrops_kingsleyae |
| ENSG00000151846 | PABPC3 | 100 | 90.110 | ENSPSIG00000003719 | PABPC1 | 100 | 90.581 | Pelodiscus_sinensis |
| ENSG00000151846 | PABPC3 | 100 | 91.366 | ENSPEMG00000014998 | Pabpc1 | 100 | 91.523 | Peromyscus_maniculatus_bairdii |
| ENSG00000151846 | PABPC3 | 100 | 77.987 | ENSPEMG00000013149 | Pabpc2 | 100 | 77.987 | Peromyscus_maniculatus_bairdii |
| ENSG00000151846 | PABPC3 | 99 | 92.021 | ENSPCIG00000000267 | - | 100 | 92.021 | Phascolarctos_cinereus |
| ENSG00000151846 | PABPC3 | 99 | 94.497 | ENSPPYG00000005219 | PABPC3 | 100 | 94.497 | Pongo_abelii |
| ENSG00000151846 | PABPC3 | 100 | 91.994 | ENSPPYG00000018790 | PABPC1 | 100 | 91.994 | Pongo_abelii |
| ENSG00000151846 | PABPC3 | 68 | 67.354 | ENSPPYG00000012833 | - | 99 | 67.354 | Pongo_abelii |
| ENSG00000151846 | PABPC3 | 100 | 90.405 | ENSPCAG00000007096 | PABPC1 | 84 | 90.405 | Procavia_capensis |
| ENSG00000151846 | PABPC3 | 100 | 85.895 | ENSPCOG00000013197 | PABPC3 | 100 | 85.895 | Propithecus_coquereli |
| ENSG00000151846 | PABPC3 | 99 | 93.019 | ENSPCOG00000020234 | PABPC1 | 100 | 93.007 | Propithecus_coquereli |
| ENSG00000151846 | PABPC3 | 100 | 91.680 | ENSPVAG00000016640 | PABPC1 | 100 | 91.680 | Pteropus_vampyrus |
| ENSG00000151846 | PABPC3 | 100 | 75.943 | ENSRNOG00000014527 | Pabpc2 | 100 | 75.943 | Rattus_norvegicus |
| ENSG00000151846 | PABPC3 | 99 | 76.352 | ENSRNOG00000017367 | Pabpc6 | 99 | 76.662 | Rattus_norvegicus |
| ENSG00000151846 | PABPC3 | 100 | 91.366 | ENSRNOG00000008639 | Pabpc1 | 100 | 91.523 | Rattus_norvegicus |
| ENSG00000151846 | PABPC3 | 99 | 88.486 | ENSRBIG00000029759 | - | 100 | 90.826 | Rhinopithecus_bieti |
| ENSG00000151846 | PABPC3 | 100 | 92.710 | ENSRBIG00000034396 | PABPC3 | 100 | 92.710 | Rhinopithecus_bieti |
| ENSG00000151846 | PABPC3 | 99 | 65.750 | ENSRBIG00000027660 | - | 100 | 65.293 | Rhinopithecus_bieti |
| ENSG00000151846 | PABPC3 | 68 | 71.963 | ENSRBIG00000000042 | - | 99 | 71.963 | Rhinopithecus_bieti |
| ENSG00000151846 | PABPC3 | 100 | 87.519 | ENSRROG00000042796 | - | 100 | 90.826 | Rhinopithecus_roxellana |
| ENSG00000151846 | PABPC3 | 66 | 64.646 | ENSRROG00000029758 | - | 93 | 64.646 | Rhinopithecus_roxellana |
| ENSG00000151846 | PABPC3 | 100 | 92.710 | ENSRROG00000036871 | PABPC3 | 100 | 92.710 | Rhinopithecus_roxellana |
| ENSG00000151846 | PABPC3 | 99 | 87.559 | ENSRROG00000032349 | - | 100 | 87.559 | Rhinopithecus_roxellana |
| ENSG00000151846 | PABPC3 | 100 | 69.731 | ENSRROG00000028723 | - | 100 | 69.731 | Rhinopithecus_roxellana |
| ENSG00000151846 | PABPC3 | 99 | 66.250 | ENSRROG00000044781 | - | 100 | 66.086 | Rhinopithecus_roxellana |
| ENSG00000151846 | PABPC3 | 100 | 86.142 | ENSSBOG00000023677 | PABPC3 | 100 | 86.142 | Saimiri_boliviensis_boliviensis |
| ENSG00000151846 | PABPC3 | 87 | 83.478 | ENSSBOG00000025499 | - | 100 | 76.147 | Saimiri_boliviensis_boliviensis |
| ENSG00000151846 | PABPC3 | 100 | 89.655 | ENSSBOG00000022216 | PABPC1 | 100 | 90.175 | Saimiri_boliviensis_boliviensis |
| ENSG00000151846 | PABPC3 | 87 | 75.000 | ENSSBOG00000026437 | - | 100 | 75.000 | Saimiri_boliviensis_boliviensis |
| ENSG00000151846 | PABPC3 | 99 | 75.160 | ENSSBOG00000020745 | - | 97 | 79.412 | Saimiri_boliviensis_boliviensis |
| ENSG00000151846 | PABPC3 | 69 | 78.733 | ENSSBOG00000021975 | - | 95 | 78.733 | Saimiri_boliviensis_boliviensis |
| ENSG00000151846 | PABPC3 | 100 | 86.142 | ENSSHAG00000003431 | - | 100 | 86.142 | Sarcophilus_harrisii |
| ENSG00000151846 | PABPC3 | 98 | 88.967 | ENSSHAG00000011267 | PABPC1 | 99 | 88.967 | Sarcophilus_harrisii |
| ENSG00000151846 | PABPC3 | 98 | 78.309 | ENSSFOG00015006617 | - | 100 | 77.743 | Scleropages_formosus |
| ENSG00000151846 | PABPC3 | 95 | 84.686 | ENSSARG00000004448 | PABPC1 | 100 | 84.686 | Sorex_araneus |
| ENSG00000151846 | PABPC3 | 100 | 90.424 | ENSSPUG00000018267 | PABPC1 | 100 | 90.424 | Sphenodon_punctatus |
| ENSG00000151846 | PABPC3 | 72 | 76.762 | ENSSPUG00000018259 | - | 98 | 76.762 | Sphenodon_punctatus |
| ENSG00000151846 | PABPC3 | 100 | 91.994 | ENSSSCG00000006063 | PABPC1 | 100 | 91.994 | Sus_scrofa |
| ENSG00000151846 | PABPC3 | 97 | 50.082 | ENSSSCG00000036199 | - | 99 | 50.082 | Sus_scrofa |
| ENSG00000151846 | PABPC3 | 100 | 89.812 | ENSTGUG00000012067 | PABPC1 | 100 | 89.969 | Taeniopygia_guttata |
| ENSG00000151846 | PABPC3 | 99 | 90.000 | ENSTBEG00000009320 | PABPC1 | 89 | 90.000 | Tupaia_belangeri |
| ENSG00000151846 | PABPC3 | 100 | 88.125 | ENSTTRG00000004689 | PABPC3 | 100 | 88.125 | Tursiops_truncatus |
| ENSG00000151846 | PABPC3 | 98 | 46.973 | ENSUAMG00000022453 | - | 99 | 47.193 | Ursus_americanus |
| ENSG00000151846 | PABPC3 | 100 | 91.994 | ENSUAMG00000024428 | PABPC1 | 100 | 91.994 | Ursus_americanus |
| ENSG00000151846 | PABPC3 | 99 | 93.110 | ENSUMAG00000017554 | PABPC1 | 100 | 93.110 | Ursus_maritimus |
| ENSG00000151846 | PABPC3 | 82 | 46.629 | ENSVPAG00000011006 | - | 98 | 46.629 | Vicugna_pacos |
| ENSG00000151846 | PABPC3 | 95 | 92.620 | ENSVPAG00000003865 | PABPC1 | 100 | 92.620 | Vicugna_pacos |
| ENSG00000151846 | PABPC3 | 99 | 93.110 | ENSVVUG00000013156 | PABPC1 | 100 | 93.110 | Vulpes_vulpes |
| ENSG00000151846 | PABPC3 | 99 | 87.579 | ENSXETG00000006635 | pabpc1 | 99 | 87.736 | Xenopus_tropicalis |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0003723 | RNA binding | 21873635. | IBA | Function |
| GO:0003730 | mRNA 3'-UTR binding | 21873635. | IBA | Function |
| GO:0005634 | nucleus | 21873635. | IBA | Component |
| GO:0005737 | cytoplasm | 21873635. | IBA | Component |
| GO:0005737 | cytoplasm | 11328870. | NAS | Component |
| GO:0005829 | cytosol | 21873635. | IBA | Component |
| GO:0005829 | cytosol | - | IDA | Component |
| GO:0008143 | poly(A) binding | 21873635. | IBA | Function |
| GO:0008143 | poly(A) binding | 11328870. | IDA | Function |
| GO:0008266 | poly(U) RNA binding | 21873635. | IBA | Function |
| GO:0010494 | cytoplasmic stress granule | 21873635. | IBA | Component |
| GO:0016071 | mRNA metabolic process | 11328870. | NAS | Process |
| GO:0070062 | extracellular exosome | 23533145. | HDA | Component |
| GO:1990904 | ribonucleoprotein complex | 21873635. | IBA | Component |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CHOL | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| DLBC | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KICH | |||||||
| KICH | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| LAML | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SARC | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| THCA | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| CESC | |||
| KIRC | |||
| MESO | |||
| THCA |
| Cancer | P-value | Q-value |
|---|---|---|
| SARC | ||
| ACC | ||
| BRCA | ||
| KIRP | ||
| PAAD | ||
| BLCA | ||
| LIHC | ||
| LGG | ||
| CHOL | ||
| LUAD | ||
| UVM | ||
| OV |