
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000425466 | MMR_HSR1 | PF01926.23 | 1.3e-05 | 1 | 1 |
| ENSP00000385432 | MMR_HSR1 | PF01926.23 | 2e-05 | 1 | 1 |
| ENSP00000404997 | MMR_HSR1 | PF01926.23 | 2e-05 | 1 | 1 |
| ENSP00000423005 | MMR_HSR1 | PF01926.23 | 0.00057 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000402673 | SAR1B-201 | 6682 | - | ENSP00000385432 | 198 (aa) | - | Q9Y6B6 |
| ENST00000508363 | SAR1B-209 | 3086 | - | - | - (aa) | - | - |
| ENST00000439578 | SAR1B-202 | 929 | - | ENSP00000404997 | 198 (aa) | - | Q9Y6B6 |
| ENST00000509937 | SAR1B-211 | 455 | - | ENSP00000424673 | 130 (aa) | - | Q9H029 |
| ENST00000505758 | SAR1B-207 | 670 | - | ENSP00000425466 | 170 (aa) | - | D6RD69 |
| ENST00000503318 | SAR1B-205 | 733 | - | ENSP00000425367 | 144 (aa) | - | D6RDB2 |
| ENST00000507419 | SAR1B-208 | 2287 | - | ENSP00000425339 | 130 (aa) | - | Q9H029 |
| ENST00000502286 | SAR1B-203 | 548 | - | ENSP00000423005 | 103 (aa) | - | D6RAA2 |
| ENST00000502539 | SAR1B-204 | 708 | - | ENSP00000426335 | 130 (aa) | - | Q9H029 |
| ENST00000504242 | SAR1B-206 | 676 | - | - | - (aa) | - | - |
| ENST00000509730 | SAR1B-210 | 506 | - | ENSP00000423197 | 118 (aa) | - | D6R9R5 |
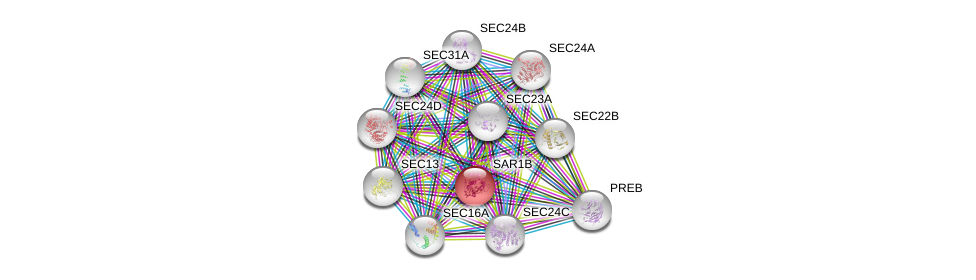
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000152700 | SAR1B | 100 | 100.000 | ENSAMEG00000017849 | SAR1B | 100 | 98.990 | Ailuropoda_melanoleuca |
| ENSG00000152700 | SAR1B | 100 | 93.204 | ENSAPLG00000007369 | SAR1A | 100 | 88.442 | Anas_platyrhynchos |
| ENSG00000152700 | SAR1B | 100 | 98.058 | ENSACAG00000001850 | SAR1B | 100 | 93.434 | Anolis_carolinensis |
| ENSG00000152700 | SAR1B | 100 | 100.000 | ENSANAG00000032734 | SAR1B | 81 | 100.000 | Aotus_nancymaae |
| ENSG00000152700 | SAR1B | 100 | 100.000 | ENSBTAG00000021226 | SAR1B | 100 | 98.485 | Bos_taurus |
| ENSG00000152700 | SAR1B | 100 | 100.000 | ENSCJAG00000004819 | SAR1B | 100 | 99.495 | Callithrix_jacchus |
| ENSG00000152700 | SAR1B | 100 | 100.000 | ENSCAFG00000031992 | SAR1B | 100 | 98.990 | Canis_familiaris |
| ENSG00000152700 | SAR1B | 100 | 100.000 | ENSCAFG00020001469 | SAR1B | 100 | 98.990 | Canis_lupus_dingo |
| ENSG00000152700 | SAR1B | 100 | 100.000 | ENSCHIG00000022509 | SAR1B | 100 | 98.485 | Capra_hircus |
| ENSG00000152700 | SAR1B | 100 | 100.000 | ENSTSYG00000011867 | SAR1B | 100 | 98.990 | Carlito_syrichta |
| ENSG00000152700 | SAR1B | 100 | 97.458 | ENSCAPG00000015216 | SAR1B | 98 | 93.299 | Cavia_aperea |
| ENSG00000152700 | SAR1B | 100 | 100.000 | ENSCPOG00000038655 | SAR1B | 100 | 97.980 | Cavia_porcellus |
| ENSG00000152700 | SAR1B | 100 | 100.000 | ENSCCAG00000037282 | SAR1B | 100 | 98.990 | Cebus_capucinus |
| ENSG00000152700 | SAR1B | 100 | 100.000 | ENSCATG00000022601 | SAR1B | 100 | 99.495 | Cercocebus_atys |
| ENSG00000152700 | SAR1B | 100 | 100.000 | ENSCLAG00000015791 | - | 100 | 98.990 | Chinchilla_lanigera |
| ENSG00000152700 | SAR1B | 100 | 100.000 | ENSCSAG00000013578 | SAR1B | 100 | 98.990 | Chlorocebus_sabaeus |
| ENSG00000152700 | SAR1B | 100 | 98.058 | ENSCPBG00000008203 | SAR1B | 100 | 96.970 | Chrysemys_picta_bellii |
| ENSG00000152700 | SAR1B | 100 | 99.029 | ENSCGRG00001016339 | SAR1B | 100 | 98.990 | Cricetulus_griseus_chok1gshd |
| ENSG00000152700 | SAR1B | 100 | 99.029 | ENSCGRG00000016600 | SAR1B | 100 | 98.990 | Cricetulus_griseus_crigri |
| ENSG00000152700 | SAR1B | 100 | 100.000 | ENSDNOG00000044440 | SAR1B | 100 | 98.990 | Dasypus_novemcinctus |
| ENSG00000152700 | SAR1B | 100 | 100.000 | ENSDORG00000023516 | Sar1b | 100 | 99.495 | Dipodomys_ordii |
| ENSG00000152700 | SAR1B | 100 | 97.980 | ENSETEG00000016178 | SAR1B | 100 | 97.980 | Echinops_telfairi |
| ENSG00000152700 | SAR1B | 100 | 100.000 | ENSEASG00005018603 | SAR1B | 100 | 99.495 | Equus_asinus_asinus |
| ENSG00000152700 | SAR1B | 100 | 100.000 | ENSECAG00000012409 | SAR1B | 100 | 99.495 | Equus_caballus |
| ENSG00000152700 | SAR1B | 100 | 100.000 | ENSFCAG00000000338 | SAR1B | 100 | 99.495 | Felis_catus |
| ENSG00000152700 | SAR1B | 100 | 94.915 | ENSFALG00000007931 | SAR1B | 99 | 95.408 | Ficedula_albicollis |
| ENSG00000152700 | SAR1B | 100 | 100.000 | ENSFDAG00000010710 | SAR1B | 100 | 98.990 | Fukomys_damarensis |
| ENSG00000152700 | SAR1B | 100 | 98.058 | ENSGALG00000038753 | SAR1B | 100 | 97.475 | Gallus_gallus |
| ENSG00000152700 | SAR1B | 100 | 98.058 | ENSGAGG00000022935 | SAR1B | 100 | 95.960 | Gopherus_agassizii |
| ENSG00000152700 | SAR1B | 100 | 99.029 | ENSHGLG00000015553 | SAR1B | 100 | 97.980 | Heterocephalus_glaber_female |
| ENSG00000152700 | SAR1B | 100 | 99.029 | ENSHGLG00100014879 | SAR1B | 100 | 97.980 | Heterocephalus_glaber_male |
| ENSG00000152700 | SAR1B | 100 | 100.000 | ENSSTOG00000011326 | - | 100 | 98.990 | Ictidomys_tridecemlineatus |
| ENSG00000152700 | SAR1B | 100 | 99.029 | ENSJJAG00000000251 | Sar1b | 100 | 98.485 | Jaculus_jaculus |
| ENSG00000152700 | SAR1B | 100 | 100.000 | ENSLAFG00000001021 | SAR1B | 100 | 98.485 | Loxodonta_africana |
| ENSG00000152700 | SAR1B | 100 | 100.000 | ENSMFAG00000002947 | SAR1B | 100 | 99.495 | Macaca_fascicularis |
| ENSG00000152700 | SAR1B | 100 | 100.000 | ENSMMUG00000004417 | SAR1B | 100 | 99.495 | Macaca_mulatta |
| ENSG00000152700 | SAR1B | 100 | 100.000 | ENSMNEG00000045379 | SAR1B | 100 | 99.495 | Macaca_nemestrina |
| ENSG00000152700 | SAR1B | 100 | 100.000 | ENSMLEG00000038596 | SAR1B | 100 | 99.495 | Mandrillus_leucophaeus |
| ENSG00000152700 | SAR1B | 100 | 98.058 | ENSMGAG00000008340 | SAR1B | 100 | 97.980 | Meleagris_gallopavo |
| ENSG00000152700 | SAR1B | 100 | 99.029 | ENSMAUG00000014677 | Sar1b | 100 | 98.990 | Mesocricetus_auratus |
| ENSG00000152700 | SAR1B | 100 | 96.923 | ENSMICG00000011638 | SAR1B | 100 | 96.465 | Microcebus_murinus |
| ENSG00000152700 | SAR1B | 100 | 99.231 | MGP_CAROLIEiJ_G0016318 | Sar1b | 100 | 98.990 | Mus_caroli |
| ENSG00000152700 | SAR1B | 100 | 99.231 | ENSMUSG00000020386 | Sar1b | 100 | 98.990 | Mus_musculus |
| ENSG00000152700 | SAR1B | 100 | 100.000 | MGP_PahariEiJ_G0017453 | Sar1b | 100 | 99.495 | Mus_pahari |
| ENSG00000152700 | SAR1B | 100 | 99.231 | MGP_SPRETEiJ_G0017162 | Sar1b | 100 | 98.990 | Mus_spretus |
| ENSG00000152700 | SAR1B | 100 | 100.000 | ENSMPUG00000010087 | SAR1B | 100 | 98.990 | Mustela_putorius_furo |
| ENSG00000152700 | SAR1B | 100 | 100.000 | ENSMLUG00000016342 | SAR1B | 100 | 98.485 | Myotis_lucifugus |
| ENSG00000152700 | SAR1B | 100 | 100.000 | ENSNGAG00000016176 | Sar1b | 100 | 98.990 | Nannospalax_galili |
| ENSG00000152700 | SAR1B | 100 | 100.000 | ENSNLEG00000008118 | SAR1B | 100 | 100.000 | Nomascus_leucogenys |
| ENSG00000152700 | SAR1B | 100 | 99.029 | ENSODEG00000009764 | SAR1B | 100 | 96.970 | Octodon_degus |
| ENSG00000152700 | SAR1B | 100 | 100.000 | ENSOCUG00000008250 | SAR1B | 100 | 98.485 | Oryctolagus_cuniculus |
| ENSG00000152700 | SAR1B | 100 | 100.000 | ENSOGAG00000013347 | SAR1B | 100 | 100.000 | Otolemur_garnettii |
| ENSG00000152700 | SAR1B | 100 | 100.000 | ENSOARG00000014231 | SAR1B | 100 | 98.485 | Ovis_aries |
| ENSG00000152700 | SAR1B | 100 | 100.000 | ENSPPAG00000033175 | SAR1B | 100 | 100.000 | Pan_paniscus |
| ENSG00000152700 | SAR1B | 100 | 100.000 | ENSPPRG00000008356 | SAR1B | 100 | 99.495 | Panthera_pardus |
| ENSG00000152700 | SAR1B | 100 | 100.000 | ENSPTIG00000001052 | SAR1B | 100 | 99.495 | Panthera_tigris_altaica |
| ENSG00000152700 | SAR1B | 100 | 100.000 | ENSPTRG00000017244 | SAR1B | 100 | 99.495 | Pan_troglodytes |
| ENSG00000152700 | SAR1B | 100 | 99.231 | ENSPANG00000005732 | SAR1B | 100 | 96.517 | Papio_anubis |
| ENSG00000152700 | SAR1B | 100 | 98.058 | ENSPSIG00000004800 | SAR1B | 100 | 96.970 | Pelodiscus_sinensis |
| ENSG00000152700 | SAR1B | 100 | 99.029 | ENSPEMG00000023610 | Sar1b | 100 | 98.990 | Peromyscus_maniculatus_bairdii |
| ENSG00000152700 | SAR1B | 100 | 98.058 | ENSPCIG00000014092 | SAR1B | 100 | 96.970 | Phascolarctos_cinereus |
| ENSG00000152700 | SAR1B | 100 | 100.000 | ENSPPYG00000015789 | SAR1B | 100 | 98.990 | Pongo_abelii |
| ENSG00000152700 | SAR1B | 100 | 100.000 | ENSPCAG00000002624 | SAR1B | 100 | 97.475 | Procavia_capensis |
| ENSG00000152700 | SAR1B | 100 | 100.000 | ENSPCOG00000025721 | - | 100 | 96.970 | Propithecus_coquereli |
| ENSG00000152700 | SAR1B | 100 | 100.000 | ENSPVAG00000012433 | SAR1B | 100 | 99.495 | Pteropus_vampyrus |
| ENSG00000152700 | SAR1B | 100 | 99.029 | ENSRNOG00000004820 | Sar1b | 100 | 98.990 | Rattus_norvegicus |
| ENSG00000152700 | SAR1B | 100 | 100.000 | ENSRBIG00000036918 | SAR1B | 100 | 99.495 | Rhinopithecus_bieti |
| ENSG00000152700 | SAR1B | 100 | 100.000 | ENSRROG00000028843 | SAR1B | 100 | 99.495 | Rhinopithecus_roxellana |
| ENSG00000152700 | SAR1B | 100 | 98.058 | ENSSHAG00000014991 | SAR1B | 100 | 96.970 | Sarcophilus_harrisii |
| ENSG00000152700 | SAR1B | 100 | 100.000 | ENSSARG00000002144 | SAR1B | 100 | 99.495 | Sorex_araneus |
| ENSG00000152700 | SAR1B | 100 | 98.058 | ENSSPUG00000009441 | SAR1B | 100 | 97.475 | Sphenodon_punctatus |
| ENSG00000152700 | SAR1B | 100 | 100.000 | ENSSSCG00000014305 | SAR1B | 100 | 98.990 | Sus_scrofa |
| ENSG00000152700 | SAR1B | 100 | 100.000 | ENSTTRG00000016864 | SAR1B | 100 | 98.990 | Tursiops_truncatus |
| ENSG00000152700 | SAR1B | 100 | 100.000 | ENSUAMG00000017620 | SAR1B | 100 | 98.485 | Ursus_americanus |
| ENSG00000152700 | SAR1B | 100 | 100.000 | ENSUMAG00000017678 | SAR1B | 100 | 98.485 | Ursus_maritimus |
| ENSG00000152700 | SAR1B | 100 | 100.000 | ENSVPAG00000001588 | - | 100 | 77.273 | Vicugna_pacos |
| ENSG00000152700 | SAR1B | 100 | 100.000 | ENSVVUG00000021294 | SAR1B | 100 | 98.990 | Vulpes_vulpes |
| ENSG00000152700 | SAR1B | 100 | 90.291 | ENSXETG00000010408 | sar1b | 99 | 84.772 | Xenopus_tropicalis |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I | - | TAS | Process |
| GO:0003400 | regulation of COPII vesicle coating | 21873635. | IBA | Process |
| GO:0003924 | GTPase activity | 21873635. | IBA | Function |
| GO:0003924 | GTPase activity | - | TAS | Function |
| GO:0005525 | GTP binding | - | IEA | Function |
| GO:0005783 | endoplasmic reticulum | 21873635. | IBA | Component |
| GO:0005789 | endoplasmic reticulum membrane | - | TAS | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0006886 | intracellular protein transport | 21873635. | IBA | Process |
| GO:0006888 | ER to Golgi vesicle-mediated transport | 21873635. | IBA | Process |
| GO:0006888 | ER to Golgi vesicle-mediated transport | - | TAS | Process |
| GO:0012507 | ER to Golgi transport vesicle membrane | - | TAS | Component |
| GO:0016050 | vesicle organization | 21873635. | IBA | Process |
| GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II | - | TAS | Process |
| GO:0030127 | COPII vesicle coat | 21873635. | IBA | Component |
| GO:0032580 | Golgi cisterna membrane | - | IEA | Component |
| GO:0046872 | metal ion binding | - | IEA | Function |
| GO:0048208 | COPII vesicle coating | - | TAS | Process |
| GO:0061024 | membrane organization | 21873635. | IBA | Process |
| GO:0070863 | positive regulation of protein exit from endoplasmic reticulum | 21873635. | IBA | Process |
| GO:0070971 | endoplasmic reticulum exit site | 21873635. | IBA | Component |