
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000283195 | zf-RanBP | PF00641.18 | 8.3e-84 | 1 | 8 |
| ENSP00000283195 | zf-RanBP | PF00641.18 | 8.3e-84 | 2 | 8 |
| ENSP00000283195 | zf-RanBP | PF00641.18 | 8.3e-84 | 3 | 8 |
| ENSP00000283195 | zf-RanBP | PF00641.18 | 8.3e-84 | 4 | 8 |
| ENSP00000283195 | zf-RanBP | PF00641.18 | 8.3e-84 | 5 | 8 |
| ENSP00000283195 | zf-RanBP | PF00641.18 | 8.3e-84 | 6 | 8 |
| ENSP00000283195 | zf-RanBP | PF00641.18 | 8.3e-84 | 7 | 8 |
| ENSP00000283195 | zf-RanBP | PF00641.18 | 8.3e-84 | 8 | 8 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000629728 | RANBP2-205 | 156 | - | ENSP00000485979 | 51 (aa) | - | F8WBP7 |
| ENST00000495924 | RANBP2-204 | 646 | - | - | - (aa) | - | - |
| ENST00000283195 | RANBP2-201 | 11711 | XM_005264002 | ENSP00000283195 | 3224 (aa) | XP_005264059 | P49792 |
| ENST00000495506 | RANBP2-203 | 544 | - | - | - (aa) | - | - |
| ENST00000425282 | RANBP2-202 | 367 | - | ENSP00000398970 | 51 (aa) | - | F8WBP7 |
| Pathway ID | Pathway Name | Source |
|---|---|---|
| hsa03013 | RNA transport | KEGG |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000153201 | rs150844750 | 2 | 108742827 | T | Attention deficit hyperactivity disorder | 29325848 | [0.15-0.35] unit decrease | 0.25 | EFO_0003888 |
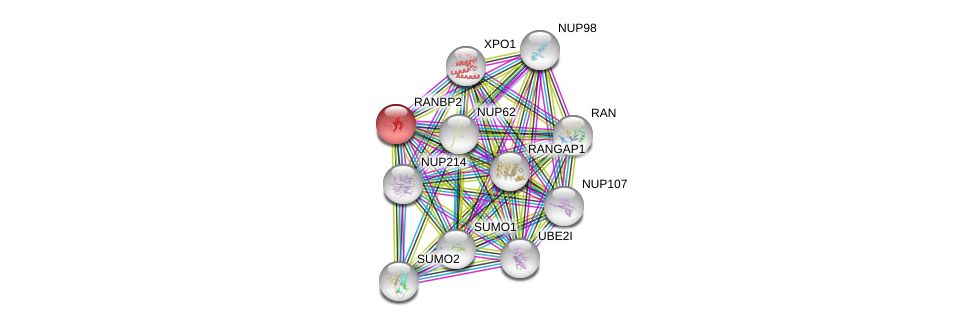
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000153201 | RANBP2 | 90 | 69.565 | ENSAPOG00000017146 | si:ch1073-55a19.2 | 100 | 54.887 | Acanthochromis_polyacanthus |
| ENSG00000153201 | RANBP2 | 99 | 87.363 | ENSAMEG00000013078 | RANBP2 | 100 | 87.395 | Ailuropoda_melanoleuca |
| ENSG00000153201 | RANBP2 | 90 | 67.391 | ENSACIG00000001203 | si:ch1073-55a19.2 | 100 | 57.143 | Amphilophus_citrinellus |
| ENSG00000153201 | RANBP2 | 90 | 69.565 | ENSAPEG00000012495 | si:ch1073-55a19.2 | 100 | 54.135 | Amphiprion_percula |
| ENSG00000153201 | RANBP2 | 90 | 67.391 | ENSATEG00000001716 | si:ch1073-55a19.2 | 100 | 53.383 | Anabas_testudineus |
| ENSG00000153201 | RANBP2 | 90 | 71.739 | ENSACAG00000002091 | RANBP2 | 100 | 64.558 | Anolis_carolinensis |
| ENSG00000153201 | RANBP2 | 100 | 93.369 | ENSANAG00000023124 | RANBP2 | 100 | 93.369 | Aotus_nancymaae |
| ENSG00000153201 | RANBP2 | 90 | 71.739 | ENSACLG00000020500 | si:ch1073-55a19.2 | 100 | 60.606 | Astatotilapia_calliptera |
| ENSG00000153201 | RANBP2 | 100 | 81.601 | ENSBTAG00000000152 | RANBP2 | 100 | 81.780 | Bos_taurus |
| ENSG00000153201 | RANBP2 | 100 | 93.957 | ENSCJAG00000001189 | RANBP2 | 100 | 93.957 | Callithrix_jacchus |
| ENSG00000153201 | RANBP2 | 100 | 83.535 | ENSCAFG00000023061 | RANBP2 | 100 | 83.658 | Canis_familiaris |
| ENSG00000153201 | RANBP2 | 100 | 89.360 | ENSCAFG00020009314 | RANBP2 | 100 | 89.483 | Canis_lupus_dingo |
| ENSG00000153201 | RANBP2 | 90 | 95.652 | ENSCHIG00000012686 | RANBP2 | 100 | 81.063 | Capra_hircus |
| ENSG00000153201 | RANBP2 | 99 | 88.655 | ENSTSYG00000001656 | RANBP2 | 100 | 88.655 | Carlito_syrichta |
| ENSG00000153201 | RANBP2 | 99 | 75.866 | ENSCAPG00000010687 | RANBP2 | 100 | 74.879 | Cavia_aperea |
| ENSG00000153201 | RANBP2 | 100 | 79.681 | ENSCPOG00000000392 | RANBP2 | 100 | 79.376 | Cavia_porcellus |
| ENSG00000153201 | RANBP2 | 100 | 93.957 | ENSCCAG00000028166 | RANBP2 | 100 | 93.957 | Cebus_capucinus |
| ENSG00000153201 | RANBP2 | 100 | 94.980 | ENSCATG00000032597 | RANBP2 | 99 | 97.142 | Cercocebus_atys |
| ENSG00000153201 | RANBP2 | 100 | 83.420 | ENSCLAG00000010533 | RANBP2 | 100 | 83.420 | Chinchilla_lanigera |
| ENSG00000153201 | RANBP2 | 100 | 96.217 | ENSCSAG00000017019 | RANBP2 | 100 | 96.217 | Chlorocebus_sabaeus |
| ENSG00000153201 | RANBP2 | 67 | 92.460 | ENSCHOG00000012659 | - | 60 | 92.460 | Choloepus_hoffmanni |
| ENSG00000153201 | RANBP2 | 100 | 69.143 | ENSCPBG00000006985 | RANBP2 | 100 | 69.459 | Chrysemys_picta_bellii |
| ENSG00000153201 | RANBP2 | 97 | 95.251 | ENSCANG00000041469 | RANBP2 | 100 | 95.251 | Colobus_angolensis_palliatus |
| ENSG00000153201 | RANBP2 | 100 | 87.041 | ENSCGRG00001000642 | Ranbp2 | 100 | 87.041 | Cricetulus_griseus_chok1gshd |
| ENSG00000153201 | RANBP2 | 99 | 87.019 | ENSCGRG00000008995 | Ranbp2 | 100 | 87.019 | Cricetulus_griseus_crigri |
| ENSG00000153201 | RANBP2 | 100 | 50.920 | ENSCSEG00000013280 | si:ch1073-55a19.2 | 100 | 57.037 | Cynoglossus_semilaevis |
| ENSG00000153201 | RANBP2 | 90 | 69.565 | ENSCVAG00000016050 | si:ch1073-55a19.2 | 100 | 49.255 | Cyprinodon_variegatus |
| ENSG00000153201 | RANBP2 | 90 | 65.217 | ENSDARG00000093125 | RANBP2 | 100 | 57.037 | Danio_rerio |
| ENSG00000153201 | RANBP2 | 100 | 84.254 | ENSDNOG00000040501 | RANBP2 | 100 | 84.371 | Dasypus_novemcinctus |
| ENSG00000153201 | RANBP2 | 100 | 89.527 | ENSDORG00000002206 | Ranbp2 | 100 | 89.527 | Dipodomys_ordii |
| ENSG00000153201 | RANBP2 | 77 | 86.657 | ENSETEG00000008891 | - | 69 | 86.501 | Echinops_telfairi |
| ENSG00000153201 | RANBP2 | 97 | 48.947 | ENSEBUG00000010057 | si:ch1073-55a19.2 | 85 | 58.088 | Eptatretus_burgeri |
| ENSG00000153201 | RANBP2 | 100 | 88.947 | ENSEASG00005012918 | RANBP2 | 100 | 88.947 | Equus_asinus_asinus |
| ENSG00000153201 | RANBP2 | 100 | 87.454 | ENSECAG00000008458 | RANBP2 | 96 | 87.392 | Equus_caballus |
| ENSG00000153201 | RANBP2 | 90 | 95.652 | ENSEEUG00000007806 | RANBP2 | 79 | 91.538 | Erinaceus_europaeus |
| ENSG00000153201 | RANBP2 | 98 | 62.381 | ENSELUG00000007763 | si:ch1073-55a19.2 | 93 | 62.381 | Esox_lucius |
| ENSG00000153201 | RANBP2 | 99 | 62.800 | ENSFALG00000003519 | RANBP2 | 99 | 62.643 | Ficedula_albicollis |
| ENSG00000153201 | RANBP2 | 100 | 80.279 | ENSFDAG00000016912 | RANBP2 | 100 | 79.976 | Fukomys_damarensis |
| ENSG00000153201 | RANBP2 | 90 | 71.739 | ENSFHEG00000009774 | si:ch1073-55a19.2 | 100 | 54.887 | Fundulus_heteroclitus |
| ENSG00000153201 | RANBP2 | 98 | 67.133 | ENSGMOG00000007071 | si:ch1073-55a19.2 | 94 | 67.133 | Gadus_morhua |
| ENSG00000153201 | RANBP2 | 90 | 67.391 | ENSGAFG00000010966 | si:ch1073-55a19.2 | 100 | 56.716 | Gambusia_affinis |
| ENSG00000153201 | RANBP2 | 90 | 71.739 | ENSGACG00000006831 | si:ch1073-55a19.2 | 100 | 54.135 | Gasterosteus_aculeatus |
| ENSG00000153201 | RANBP2 | 90 | 82.609 | ENSGAGG00000025039 | RANBP2 | 99 | 69.168 | Gopherus_agassizii |
| ENSG00000153201 | RANBP2 | 90 | 71.739 | ENSHBUG00000000053 | si:ch1073-55a19.2 | 100 | 60.606 | Haplochromis_burtoni |
| ENSG00000153201 | RANBP2 | 100 | 85.317 | ENSHGLG00000018887 | RANBP2 | 100 | 85.317 | Heterocephalus_glaber_female |
| ENSG00000153201 | RANBP2 | 100 | 82.971 | ENSHGLG00100007365 | RANBP2 | 100 | 83.590 | Heterocephalus_glaber_male |
| ENSG00000153201 | RANBP2 | 90 | 65.217 | ENSHCOG00000019596 | si:ch1073-55a19.2 | 100 | 51.002 | Hippocampus_comes |
| ENSG00000153201 | RANBP2 | 90 | 73.913 | ENSIPUG00000019614 | si:ch1073-55a19.2 | 99 | 52.709 | Ictalurus_punctatus |
| ENSG00000153201 | RANBP2 | 100 | 86.061 | ENSSTOG00000019518 | RANBP2 | 100 | 83.938 | Ictidomys_tridecemlineatus |
| ENSG00000153201 | RANBP2 | 100 | 86.507 | ENSJJAG00000021292 | Ranbp2 | 100 | 86.507 | Jaculus_jaculus |
| ENSG00000153201 | RANBP2 | 90 | 69.565 | ENSKMAG00000001158 | si:ch1073-55a19.2 | 100 | 54.887 | Kryptolebias_marmoratus |
| ENSG00000153201 | RANBP2 | 90 | 60.870 | ENSLBEG00000027979 | si:ch1073-55a19.2 | 99 | 55.556 | Labrus_bergylta |
| ENSG00000153201 | RANBP2 | 100 | 58.194 | ENSLACG00000009728 | RANBP2 | 100 | 58.119 | Latimeria_chalumnae |
| ENSG00000153201 | RANBP2 | 90 | 67.391 | ENSLOCG00000010333 | si:ch1073-55a19.2 | 100 | 55.948 | Lepisosteus_oculatus |
| ENSG00000153201 | RANBP2 | 90 | 95.652 | ENSLAFG00000000740 | RANBP2 | 100 | 84.322 | Loxodonta_africana |
| ENSG00000153201 | RANBP2 | 100 | 97.116 | ENSMFAG00000029029 | RANBP2 | 100 | 97.116 | Macaca_fascicularis |
| ENSG00000153201 | RANBP2 | 100 | 97.085 | ENSMMUG00000021019 | RANBP2 | 100 | 97.085 | Macaca_mulatta |
| ENSG00000153201 | RANBP2 | 100 | 97.209 | ENSMNEG00000043706 | RANBP2 | 100 | 97.209 | Macaca_nemestrina |
| ENSG00000153201 | RANBP2 | 99 | 96.420 | ENSMLEG00000037350 | RANBP2 | 100 | 97.429 | Mandrillus_leucophaeus |
| ENSG00000153201 | RANBP2 | 90 | 69.565 | ENSMAMG00000017900 | si:ch1073-55a19.2 | 100 | 54.135 | Mastacembelus_armatus |
| ENSG00000153201 | RANBP2 | 90 | 71.739 | ENSMZEG00005010956 | si:ch1073-55a19.2 | 100 | 60.606 | Maylandia_zebra |
| ENSG00000153201 | RANBP2 | 99 | 63.797 | ENSMGAG00000014831 | - | 99 | 63.400 | Meleagris_gallopavo |
| ENSG00000153201 | RANBP2 | 100 | 86.682 | ENSMAUG00000013313 | Ranbp2 | 100 | 86.682 | Mesocricetus_auratus |
| ENSG00000153201 | RANBP2 | 100 | 90.653 | ENSMICG00000011864 | RANBP2 | 100 | 90.622 | Microcebus_murinus |
| ENSG00000153201 | RANBP2 | 100 | 87.316 | ENSMOCG00000012516 | Ranbp2 | 100 | 87.316 | Microtus_ochrogaster |
| ENSG00000153201 | RANBP2 | 90 | 65.217 | ENSMMOG00000005743 | si:ch1073-55a19.2 | 100 | 54.426 | Mola_mola |
| ENSG00000153201 | RANBP2 | 98 | 84.267 | ENSMODG00000002914 | RANBP2 | 100 | 84.267 | Monodelphis_domestica |
| ENSG00000153201 | RANBP2 | 90 | 69.565 | ENSMALG00000004359 | si:ch1073-55a19.2 | 100 | 52.593 | Monopterus_albus |
| ENSG00000153201 | RANBP2 | 90 | 95.652 | ENSMUSG00000003226 | Ranbp2 | 100 | 84.329 | Mus_musculus |
| ENSG00000153201 | RANBP2 | 90 | 95.652 | MGP_PahariEiJ_G0030751 | Ranbp2 | 100 | 84.447 | Mus_pahari |
| ENSG00000153201 | RANBP2 | 90 | 95.652 | MGP_SPRETEiJ_G0016147 | Ranbp2 | 100 | 84.506 | Mus_spretus |
| ENSG00000153201 | RANBP2 | 100 | 88.640 | ENSMPUG00000010754 | RANBP2 | 100 | 88.640 | Mustela_putorius_furo |
| ENSG00000153201 | RANBP2 | 99 | 86.053 | ENSMLUG00000005555 | RANBP2 | 100 | 86.053 | Myotis_lucifugus |
| ENSG00000153201 | RANBP2 | 100 | 88.521 | ENSNGAG00000016154 | Ranbp2 | 100 | 88.521 | Nannospalax_galili |
| ENSG00000153201 | RANBP2 | 90 | 71.739 | ENSNBRG00000004587 | si:ch1073-55a19.2 | 100 | 60.606 | Neolamprologus_brichardi |
| ENSG00000153201 | RANBP2 | 99 | 94.580 | ENSNLEG00000007501 | RANBP2 | 99 | 94.580 | Nomascus_leucogenys |
| ENSG00000153201 | RANBP2 | 92 | 86.016 | ENSOPRG00000009720 | - | 94 | 86.016 | Ochotona_princeps |
| ENSG00000153201 | RANBP2 | 100 | 81.639 | ENSODEG00000013212 | RANBP2 | 100 | 81.987 | Octodon_degus |
| ENSG00000153201 | RANBP2 | 90 | 71.739 | ENSONIG00000004346 | si:ch1073-55a19.2 | 100 | 59.398 | Oreochromis_niloticus |
| ENSG00000153201 | RANBP2 | 80 | 69.894 | ENSOANG00000004894 | RANBP2 | 100 | 70.061 | Ornithorhynchus_anatinus |
| ENSG00000153201 | RANBP2 | 90 | 95.652 | ENSOCUG00000001165 | RANBP2 | 100 | 83.525 | Oryctolagus_cuniculus |
| ENSG00000153201 | RANBP2 | 100 | 50.313 | ENSORLG00000015523 | si:ch1073-55a19.2 | 100 | 50.166 | Oryzias_latipes |
| ENSG00000153201 | RANBP2 | 90 | 63.043 | ENSORLG00020003797 | si:ch1073-55a19.2 | 100 | 56.391 | Oryzias_latipes_hni |
| ENSG00000153201 | RANBP2 | 100 | 51.701 | ENSORLG00015004994 | si:ch1073-55a19.2 | 100 | 56.391 | Oryzias_latipes_hsok |
| ENSG00000153201 | RANBP2 | 90 | 69.565 | ENSOMEG00000022969 | si:ch1073-55a19.2 | 100 | 54.887 | Oryzias_melastigma |
| ENSG00000153201 | RANBP2 | 100 | 85.135 | ENSOGAG00000028976 | RANBP2 | 100 | 86.069 | Otolemur_garnettii |
| ENSG00000153201 | RANBP2 | 99 | 81.329 | ENSOARG00000000558 | RANBP2 | 100 | 81.390 | Ovis_aries |
| ENSG00000153201 | RANBP2 | 100 | 95.229 | ENSPANG00000000990 | RANBP2 | 100 | 95.229 | Papio_anubis |
| ENSG00000153201 | RANBP2 | 93 | 52.518 | ENSPKIG00000021054 | - | 99 | 52.672 | Paramormyrops_kingsleyae |
| ENSG00000153201 | RANBP2 | 90 | 67.391 | ENSPKIG00000010971 | RANBP2 | 100 | 55.556 | Paramormyrops_kingsleyae |
| ENSG00000153201 | RANBP2 | 99 | 68.694 | ENSPSIG00000007182 | RANBP2 | 99 | 68.934 | Pelodiscus_sinensis |
| ENSG00000153201 | RANBP2 | 94 | 87.895 | ENSPEMG00000021565 | Ranbp2 | 97 | 87.895 | Peromyscus_maniculatus_bairdii |
| ENSG00000153201 | RANBP2 | 85 | 55.797 | ENSPMAG00000007195 | si:ch1073-55a19.2 | 100 | 61.314 | Petromyzon_marinus |
| ENSG00000153201 | RANBP2 | 90 | 91.304 | ENSPCIG00000013912 | RANBP2 | 100 | 77.743 | Phascolarctos_cinereus |
| ENSG00000153201 | RANBP2 | 90 | 63.043 | ENSPFOG00000007377 | si:ch1073-55a19.2 | 100 | 49.216 | Poecilia_formosa |
| ENSG00000153201 | RANBP2 | 90 | 63.043 | ENSPLAG00000012373 | si:ch1073-55a19.2 | 100 | 55.970 | Poecilia_latipinna |
| ENSG00000153201 | RANBP2 | 90 | 63.043 | ENSPMEG00000005561 | si:ch1073-55a19.2 | 100 | 49.065 | Poecilia_mexicana |
| ENSG00000153201 | RANBP2 | 90 | 67.391 | ENSPREG00000013331 | si:ch1073-55a19.2 | 99 | 66.923 | Poecilia_reticulata |
| ENSG00000153201 | RANBP2 | 73 | 98.345 | ENSPPYG00000012139 | - | 100 | 98.345 | Pongo_abelii |
| ENSG00000153201 | RANBP2 | 55 | 90.826 | ENSPPYG00000029793 | - | 99 | 90.826 | Pongo_abelii |
| ENSG00000153201 | RANBP2 | 97 | 80.038 | ENSPCAG00000000957 | RANBP2 | 100 | 80.038 | Procavia_capensis |
| ENSG00000153201 | RANBP2 | 100 | 86.103 | ENSPCOG00000008879 | RANBP2 | 100 | 89.628 | Propithecus_coquereli |
| ENSG00000153201 | RANBP2 | 98 | 89.240 | ENSPVAG00000003883 | RANBP2 | 94 | 89.240 | Pteropus_vampyrus |
| ENSG00000153201 | RANBP2 | 90 | 71.739 | ENSPNYG00000015971 | si:ch1073-55a19.2 | 100 | 60.606 | Pundamilia_nyererei |
| ENSG00000153201 | RANBP2 | 90 | 71.739 | ENSPNAG00000026129 | si:ch1073-55a19.2 | 100 | 51.526 | Pygocentrus_nattereri |
| ENSG00000153201 | RANBP2 | 100 | 86.568 | ENSRNOG00000000796 | Ranbp2 | 100 | 86.568 | Rattus_norvegicus |
| ENSG00000153201 | RANBP2 | 100 | 94.672 | ENSRBIG00000043515 | RANBP2 | 100 | 94.672 | Rhinopithecus_bieti |
| ENSG00000153201 | RANBP2 | 100 | 94.574 | ENSRROG00000026113 | RANBP2 | 100 | 94.574 | Rhinopithecus_roxellana |
| ENSG00000153201 | RANBP2 | 100 | 93.317 | ENSSBOG00000035336 | RANBP2 | 100 | 93.317 | Saimiri_boliviensis_boliviensis |
| ENSG00000153201 | RANBP2 | 99 | 77.641 | ENSSHAG00000004676 | RANBP2 | 99 | 77.641 | Sarcophilus_harrisii |
| ENSG00000153201 | RANBP2 | 90 | 67.391 | ENSSFOG00015003943 | - | 99 | 53.998 | Scleropages_formosus |
| ENSG00000153201 | RANBP2 | 90 | 65.217 | ENSSFOG00015023201 | RANBP2 | 100 | 57.037 | Scleropages_formosus |
| ENSG00000153201 | RANBP2 | 90 | 69.565 | ENSSMAG00000007331 | si:ch1073-55a19.2 | 100 | 54.676 | Scophthalmus_maximus |
| ENSG00000153201 | RANBP2 | 90 | 69.565 | ENSSDUG00000013160 | si:ch1073-55a19.2 | 100 | 54.135 | Seriola_dumerili |
| ENSG00000153201 | RANBP2 | 90 | 69.565 | ENSSLDG00000023392 | si:ch1073-55a19.2 | 100 | 54.676 | Seriola_lalandi_dorsalis |
| ENSG00000153201 | RANBP2 | 89 | 87.975 | ENSSARG00000005595 | RANBP2 | 77 | 87.975 | Sorex_araneus |
| ENSG00000153201 | RANBP2 | 90 | 69.565 | ENSSPAG00000008000 | si:ch1073-55a19.2 | 100 | 54.135 | Stegastes_partitus |
| ENSG00000153201 | RANBP2 | 90 | 95.652 | ENSSSCG00000008136 | RANBP2 | 98 | 84.711 | Sus_scrofa |
| ENSG00000153201 | RANBP2 | 98 | 69.840 | ENSTGUG00000009571 | RANBP2 | 100 | 69.919 | Taeniopygia_guttata |
| ENSG00000153201 | RANBP2 | 95 | 90.210 | ENSTTRG00000007400 | RANBP2 | 96 | 90.210 | Tursiops_truncatus |
| ENSG00000153201 | RANBP2 | 94 | 89.667 | ENSUAMG00000020025 | RANBP2 | 98 | 89.667 | Ursus_americanus |
| ENSG00000153201 | RANBP2 | 96 | 81.542 | ENSVPAG00000006676 | RANBP2 | 94 | 81.735 | Vicugna_pacos |
| ENSG00000153201 | RANBP2 | 100 | 89.338 | ENSVVUG00000014899 | RANBP2 | 100 | 89.338 | Vulpes_vulpes |
| ENSG00000153201 | RANBP2 | 90 | 71.739 | ENSXETG00000016006 | ranbp2 | 99 | 56.061 | Xenopus_tropicalis |
| ENSG00000153201 | RANBP2 | 99 | 52.941 | ENSXCOG00000000752 | si:ch1073-55a19.2 | 99 | 52.941 | Xiphophorus_couchianus |
| ENSG00000153201 | RANBP2 | 90 | 69.565 | ENSXMAG00000003519 | si:ch1073-55a19.2 | 100 | 52.941 | Xiphophorus_maculatus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000413 | protein peptidyl-prolyl isomerization | - | IEA | Process |
| GO:0001975 | response to amphetamine | - | IEA | Process |
| GO:0003723 | RNA binding | - | IEA | Function |
| GO:0003755 | peptidyl-prolyl cis-trans isomerase activity | 20676357. | IDA | Function |
| GO:0005096 | GTPase activator activity | 21873635. | IBA | Function |
| GO:0005515 | protein binding | 10078529.14729961.16314522.16332688.20386726.21988832.22155184.23414517.24855949.26272610. | IPI | Function |
| GO:0005634 | nucleus | 21873635. | IBA | Component |
| GO:0005635 | nuclear envelope | 20386726. | IDA | Component |
| GO:0005635 | nuclear envelope | - | TAS | Component |
| GO:0005642 | annulate lamellae | 20386726. | IDA | Component |
| GO:0005643 | nuclear pore | 21873635. | IBA | Component |
| GO:0005643 | nuclear pore | 17098863.20386726. | IDA | Component |
| GO:0005737 | cytoplasm | 21873635. | IBA | Component |
| GO:0005739 | mitochondrion | - | IEA | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0006110 | regulation of glycolytic process | - | TAS | Process |
| GO:0006111 | regulation of gluconeogenesis | - | IEA | Process |
| GO:0006406 | mRNA export from nucleus | - | TAS | Process |
| GO:0006409 | tRNA export from nucleus | - | TAS | Process |
| GO:0006457 | protein folding | - | IEA | Process |
| GO:0006607 | NLS-bearing protein import into nucleus | 21873635. | IBA | Process |
| GO:0006607 | NLS-bearing protein import into nucleus | 7603572. | IMP | Process |
| GO:0008536 | Ran GTPase binding | 21873635. | IBA | Function |
| GO:0008536 | Ran GTPase binding | 7603572. | IPI | Function |
| GO:0016018 | cyclosporin A binding | 20676357. | IDA | Function |
| GO:0016020 | membrane | 19946888. | HDA | Component |
| GO:0016032 | viral process | - | TAS | Process |
| GO:0016925 | protein sumoylation | 17264123.22155184. | IDA | Process |
| GO:0016925 | protein sumoylation | - | IEA | Process |
| GO:0016925 | protein sumoylation | - | TAS | Process |
| GO:0019083 | viral transcription | - | TAS | Process |
| GO:0019789 | SUMO transferase activity | 11792325.15378033.15608651.16688858. | EXP | Function |
| GO:0031965 | nuclear membrane | 11839768. | IDA | Component |
| GO:0033133 | positive regulation of glucokinase activity | - | IEA | Process |
| GO:0042405 | nuclear inclusion body | 11839768. | IDA | Component |
| GO:0043547 | positive regulation of GTPase activity | - | IEA | Process |
| GO:0043657 | host cell | - | IEA | Component |
| GO:0044614 | nuclear pore cytoplasmic filaments | 7603572. | IDA | Component |
| GO:0044615 | nuclear pore nuclear basket | 11839768. | IDA | Component |
| GO:0044877 | protein-containing complex binding | - | IEA | Function |
| GO:0046872 | metal ion binding | - | IEA | Function |
| GO:0051642 | centrosome localization | 20386726. | IMP | Process |
| GO:0060964 | regulation of gene silencing by miRNA | - | TAS | Process |
| GO:0075733 | intracellular transport of virus | - | TAS | Process |
| GO:1900034 | regulation of cellular response to heat | - | TAS | Process |
| GO:1990723 | cytoplasmic periphery of the nuclear pore complex | - | IEA | Component |