
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000358658 | ResIII | PF04851.15 | 5.9e-06 | 1 | 1 |
| ENSP00000350028 | ResIII | PF04851.15 | 6.5e-06 | 1 | 1 |
| ENSP00000358659 | ResIII | PF04851.15 | 6.5e-06 | 1 | 1 |
| ENSP00000399797 | ResIII | PF04851.15 | 6.5e-06 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000488160 | MOV10-213 | 896 | - | - | - (aa) | - | - |
| ENST00000465579 | MOV10-205 | 610 | - | - | - (aa) | - | - |
| ENST00000479858 | MOV10-209 | 400 | - | - | - (aa) | - | - |
| ENST00000490413 | MOV10-214 | 1370 | - | - | - (aa) | - | - |
| ENST00000481711 | MOV10-210 | 379 | - | - | - (aa) | - | - |
| ENST00000486416 | MOV10-212 | 347 | - | - | - (aa) | - | - |
| ENST00000369644 | MOV10-202 | 4100 | XM_005270869 | ENSP00000358658 | 947 (aa) | XP_005270926 | Q5JR04 |
| ENST00000496577 | MOV10-217 | 4312 | - | - | - (aa) | - | - |
| ENST00000475429 | MOV10-208 | 582 | - | - | - (aa) | - | - |
| ENST00000369645 | MOV10-203 | 3552 | XM_017001319 | ENSP00000358659 | 1003 (aa) | XP_016856808 | Q9HCE1 |
| ENST00000413052 | MOV10-204 | 3641 | - | ENSP00000399797 | 1003 (aa) | - | Q9HCE1 |
| ENST00000468624 | MOV10-206 | 4689 | - | - | - (aa) | - | - |
| ENST00000357443 | MOV10-201 | 3383 | XM_024447132 | ENSP00000350028 | 1003 (aa) | XP_024302900 | Q9HCE1 |
| ENST00000495374 | MOV10-216 | 705 | - | - | - (aa) | - | - |
| ENST00000471160 | MOV10-207 | 493 | - | - | - (aa) | - | - |
| ENST00000494319 | MOV10-215 | 720 | - | - | - (aa) | - | - |
| ENST00000482545 | MOV10-211 | 380 | - | - | - (aa) | - | - |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000155363 | Blood Pressure | 1E-9 | 21909115 |
| ENSG00000155363 | Hypertension | 3E-7 | 21909115 |
| ENSG00000155363 | Blood Pressure | 1E-9 | 21909115 |
| ENSG00000155363 | Hip | 9E-7 | 25673412 |
| ENSG00000155363 | Body Mass Index | 9E-7 | 25673412 |
| ENSG00000155363 | Hip | 6E-9 | 25673412 |
| ENSG00000155363 | Body Mass Index | 6E-9 | 25673412 |
| ENSG00000155363 | Arterial Pressure | 8E-6 | 21909110 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000155363 | rs12129649 | 1 | 112688881 | T | Diastolic blood pressure | 28739976 | [0.44-0.83] unit increase | 0.638 | EFO_0006336 |
| ENSG00000155363 | rs2932538 | 1 | 112673921 | G | Diastolic blood pressure | 28739976 | [0.18-0.37] unit increase | 0.278 | EFO_0006336 |
| ENSG00000155363 | rs12129649 | 1 | 112688881 | T | Diastolic blood pressure | 28739976 | [0.43-0.67] unit increase | 0.548 | EFO_0006336 |
| ENSG00000155363 | rs2932538 | 1 | 112673921 | G | Systolic blood pressure | 28739976 | [0.27-0.58] unit increase | 0.424 | EFO_0006335 |
| ENSG00000155363 | rs2999159 | 1 | 112688136 | A | Systolic blood pressure x alcohol consumption interaction (2df test) | 29912962 | EFO_0006335|EFO_0004329 | ||
| ENSG00000155363 | rs2999159 | 1 | 112688136 | A | Diastolic blood pressure x alcohol consumption interaction (2df test) | 29912962 | EFO_0006336|EFO_0004329 | ||
| ENSG00000155363 | rs2999159 | 1 | 112688136 | A | Mean arterial pressure x alcohol consumption interaction (2df test) | 29912962 | EFO_0006340|EFO_0004329 | ||
| ENSG00000155363 | rs2932538 | 1 | 112673921 | G | Blood pressure | 21909110 | [0.14-0.36] mmHg increase | 0.252 | EFO_0006340 |
| ENSG00000155363 | rs2932538 | 1 | 112673921 | G | Diastolic blood pressure | 21909115 | [NR] mmHg increase | 0.24 | EFO_0006336 |
| ENSG00000155363 | rs2932538 | 1 | 112673921 | G | Hypertension | 21909115 | [NR] unit increase | 0.049 | EFO_0000537 |
| ENSG00000155363 | rs2932538 | 1 | 112673921 | G | Systolic blood pressure | 21909115 | [NR] mmHg increase | 0.388 | EFO_0006335 |
| ENSG00000155363 | rs3748656 | 1 | 112694059 | T | Hip circumference adjusted for BMI | 25673412 | [0.016-0.033] unit decrease | 0.0244 | EFO_0008039 |
| ENSG00000155363 | rs3748656 | 1 | 112694059 | T | Hip circumference adjusted for BMI | 25673412 | [0.016-0.037] unit decrease | 0.0267 | EFO_0008039 |
| ENSG00000155363 | rs2999158 | 1 | 112696856 | ? | Lung function (FVC) | 30595370 | EFO_0004312 | ||
| ENSG00000155363 | rs2932538 | 1 | 112673921 | A | Diastolic blood pressure | 27841878 | unit decrease | 0.185 | EFO_0006336 |
| ENSG00000155363 | rs2932538 | 1 | 112673921 | A | Systolic blood pressure | 27841878 | unit decrease | 0.227 | EFO_0006335 |
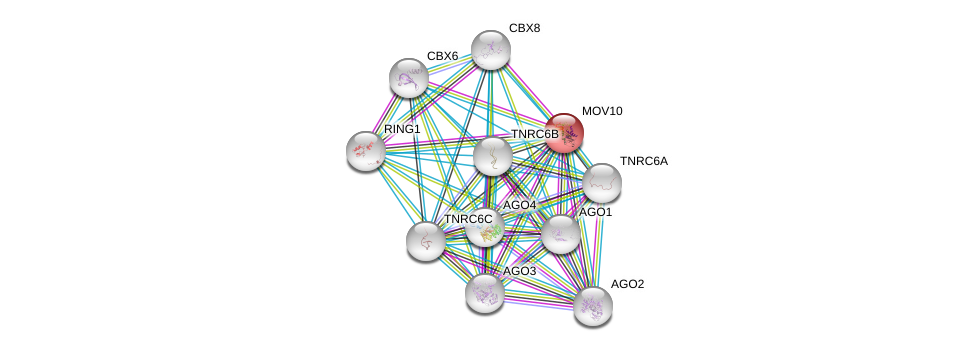
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000155363 | MOV10 | 86 | 40.000 | ENSAPOG00000003017 | mov10b.1 | 99 | 40.000 | Acanthochromis_polyacanthus |
| ENSG00000155363 | MOV10 | 100 | 95.565 | ENSAMEG00000003505 | MOV10 | 98 | 95.414 | Ailuropoda_melanoleuca |
| ENSG00000155363 | MOV10 | 95 | 45.564 | ENSACIG00000012386 | mov10b.1 | 89 | 45.564 | Amphilophus_citrinellus |
| ENSG00000155363 | MOV10 | 99 | 43.973 | ENSATEG00000023014 | mov10b.1 | 99 | 43.877 | Anabas_testudineus |
| ENSG00000155363 | MOV10 | 88 | 51.483 | ENSAPLG00000015955 | MOV10 | 97 | 49.073 | Anas_platyrhynchos |
| ENSG00000155363 | MOV10 | 99 | 46.765 | ENSACAG00000017938 | MOV10 | 99 | 46.765 | Anolis_carolinensis |
| ENSG00000155363 | MOV10 | 100 | 98.405 | ENSANAG00000034430 | MOV10 | 100 | 98.405 | Aotus_nancymaae |
| ENSG00000155363 | MOV10 | 95 | 45.235 | ENSACLG00000007569 | mov10b.1 | 97 | 43.012 | Astatotilapia_calliptera |
| ENSG00000155363 | MOV10 | 90 | 46.379 | ENSAMXG00000000035 | mov10b.1 | 96 | 46.379 | Astyanax_mexicanus |
| ENSG00000155363 | MOV10 | 100 | 92.503 | ENSBTAG00000014297 | MOV10 | 100 | 92.124 | Bos_taurus |
| ENSG00000155363 | MOV10 | 100 | 98.604 | ENSCJAG00000013592 | MOV10 | 100 | 98.604 | Callithrix_jacchus |
| ENSG00000155363 | MOV10 | 100 | 95.037 | ENSCAFG00000013454 | MOV10 | 100 | 94.816 | Canis_familiaris |
| ENSG00000155363 | MOV10 | 100 | 95.037 | ENSCAFG00020008556 | MOV10 | 100 | 94.816 | Canis_lupus_dingo |
| ENSG00000155363 | MOV10 | 100 | 92.080 | ENSCHIG00000022479 | MOV10 | 100 | 91.725 | Capra_hircus |
| ENSG00000155363 | MOV10 | 100 | 95.037 | ENSTSYG00000034936 | MOV10 | 100 | 95.037 | Carlito_syrichta |
| ENSG00000155363 | MOV10 | 93 | 80.181 | ENSCAPG00000008094 | MOV10 | 97 | 80.201 | Cavia_aperea |
| ENSG00000155363 | MOV10 | 100 | 88.701 | ENSCPOG00000014537 | MOV10 | 100 | 88.235 | Cavia_porcellus |
| ENSG00000155363 | MOV10 | 100 | 98.405 | ENSCCAG00000034901 | MOV10 | 100 | 98.405 | Cebus_capucinus |
| ENSG00000155363 | MOV10 | 100 | 99.003 | ENSCATG00000036718 | MOV10 | 100 | 99.003 | Cercocebus_atys |
| ENSG00000155363 | MOV10 | 100 | 89.757 | ENSCLAG00000008493 | MOV10 | 100 | 89.332 | Chinchilla_lanigera |
| ENSG00000155363 | MOV10 | 100 | 99.003 | ENSCSAG00000001600 | MOV10 | 100 | 99.003 | Chlorocebus_sabaeus |
| ENSG00000155363 | MOV10 | 99 | 50.934 | ENSCPBG00000018937 | MOV10 | 99 | 51.327 | Chrysemys_picta_bellii |
| ENSG00000155363 | MOV10 | 76 | 44.536 | ENSCING00000023723 | - | 94 | 44.536 | Ciona_intestinalis |
| ENSG00000155363 | MOV10 | 100 | 99.202 | ENSCANG00000039385 | MOV10 | 100 | 99.202 | Colobus_angolensis_palliatus |
| ENSG00000155363 | MOV10 | 100 | 92.089 | ENSCGRG00001010914 | Mov10 | 100 | 91.534 | Cricetulus_griseus_chok1gshd |
| ENSG00000155363 | MOV10 | 100 | 92.089 | ENSCGRG00000003973 | Mov10 | 99 | 91.979 | Cricetulus_griseus_crigri |
| ENSG00000155363 | MOV10 | 94 | 41.969 | ENSCVAG00000012443 | - | 96 | 41.969 | Cyprinodon_variegatus |
| ENSG00000155363 | MOV10 | 89 | 45.476 | ENSCVAG00000002355 | mov10a | 90 | 45.476 | Cyprinodon_variegatus |
| ENSG00000155363 | MOV10 | 94 | 43.838 | ENSDARG00000061177 | mov10b.1 | 95 | 43.838 | Danio_rerio |
| ENSG00000155363 | MOV10 | 99 | 43.212 | ENSDARG00000015829 | mov10a | 99 | 43.436 | Danio_rerio |
| ENSG00000155363 | MOV10 | 100 | 92.291 | ENSDNOG00000007631 | MOV10 | 93 | 89.296 | Dasypus_novemcinctus |
| ENSG00000155363 | MOV10 | 100 | 92.503 | ENSDORG00000000673 | Mov10 | 100 | 92.124 | Dipodomys_ordii |
| ENSG00000155363 | MOV10 | 100 | 85.579 | ENSETEG00000002395 | MOV10 | 100 | 85.388 | Echinops_telfairi |
| ENSG00000155363 | MOV10 | 100 | 96.515 | ENSEASG00005005486 | MOV10 | 100 | 96.510 | Equus_asinus_asinus |
| ENSG00000155363 | MOV10 | 100 | 96.621 | ENSECAG00000011530 | MOV10 | 100 | 96.621 | Equus_caballus |
| ENSG00000155363 | MOV10 | 81 | 53.077 | ENSELUG00000000566 | mov10b.1 | 99 | 49.826 | Esox_lucius |
| ENSG00000155363 | MOV10 | 100 | 96.304 | ENSFCAG00000013305 | MOV10 | 100 | 96.304 | Felis_catus |
| ENSG00000155363 | MOV10 | 95 | 49.066 | ENSFALG00000000722 | MOV10 | 98 | 49.274 | Ficedula_albicollis |
| ENSG00000155363 | MOV10 | 100 | 89.863 | ENSFDAG00000007746 | MOV10 | 100 | 89.133 | Fukomys_damarensis |
| ENSG00000155363 | MOV10 | 98 | 44.759 | ENSFHEG00000005435 | - | 99 | 42.898 | Fundulus_heteroclitus |
| ENSG00000155363 | MOV10 | 89 | 35.772 | ENSFHEG00000011343 | - | 90 | 35.772 | Fundulus_heteroclitus |
| ENSG00000155363 | MOV10 | 89 | 55.615 | ENSGAFG00000007675 | mov10b.1 | 96 | 55.615 | Gambusia_affinis |
| ENSG00000155363 | MOV10 | 96 | 44.408 | ENSGAFG00000000774 | mov10a | 90 | 44.408 | Gambusia_affinis |
| ENSG00000155363 | MOV10 | 93 | 52.125 | ENSGAGG00000018740 | MOV10 | 100 | 51.371 | Gopherus_agassizii |
| ENSG00000155363 | MOV10 | 100 | 100.000 | ENSGGOG00000002731 | MOV10 | 100 | 100.000 | Gorilla_gorilla |
| ENSG00000155363 | MOV10 | 95 | 43.187 | ENSHBUG00000005915 | mov10b.1 | 97 | 40.792 | Haplochromis_burtoni |
| ENSG00000155363 | MOV10 | 100 | 90.074 | ENSHGLG00000004970 | MOV10 | 100 | 89.432 | Heterocephalus_glaber_female |
| ENSG00000155363 | MOV10 | 100 | 90.074 | ENSHGLG00100004320 | MOV10 | 99 | 90.074 | Heterocephalus_glaber_male |
| ENSG00000155363 | MOV10 | 89 | 46.799 | ENSHCOG00000019421 | mov10b.1 | 93 | 46.915 | Hippocampus_comes |
| ENSG00000155363 | MOV10 | 99 | 43.254 | ENSIPUG00000023578 | mov10b.1 | 99 | 43.254 | Ictalurus_punctatus |
| ENSG00000155363 | MOV10 | 99 | 42.967 | ENSIPUG00000004768 | mov10b.1 | 99 | 42.967 | Ictalurus_punctatus |
| ENSG00000155363 | MOV10 | 100 | 96.211 | ENSSTOG00000015963 | - | 100 | 96.211 | Ictidomys_tridecemlineatus |
| ENSG00000155363 | MOV10 | 100 | 91.043 | ENSJJAG00000016562 | Mov10 | 100 | 90.945 | Jaculus_jaculus |
| ENSG00000155363 | MOV10 | 100 | 44.623 | ENSKMAG00000020712 | MOV10 | 97 | 44.575 | Kryptolebias_marmoratus |
| ENSG00000155363 | MOV10 | 65 | 38.141 | ENSKMAG00000013343 | - | 84 | 38.141 | Kryptolebias_marmoratus |
| ENSG00000155363 | MOV10 | 80 | 37.630 | ENSLBEG00000009539 | - | 91 | 37.630 | Labrus_bergylta |
| ENSG00000155363 | MOV10 | 99 | 43.732 | ENSLBEG00000009630 | - | 99 | 43.732 | Labrus_bergylta |
| ENSG00000155363 | MOV10 | 100 | 43.223 | ENSLACG00000017920 | MOV10 | 99 | 43.223 | Latimeria_chalumnae |
| ENSG00000155363 | MOV10 | 100 | 95.987 | ENSLAFG00000000239 | MOV10 | 99 | 95.386 | Loxodonta_africana |
| ENSG00000155363 | MOV10 | 100 | 99.003 | ENSMFAG00000002283 | MOV10 | 100 | 99.003 | Macaca_fascicularis |
| ENSG00000155363 | MOV10 | 100 | 99.003 | ENSMNEG00000040033 | MOV10 | 100 | 99.003 | Macaca_nemestrina |
| ENSG00000155363 | MOV10 | 100 | 99.003 | ENSMLEG00000001940 | MOV10 | 100 | 99.003 | Mandrillus_leucophaeus |
| ENSG00000155363 | MOV10 | 95 | 44.676 | ENSMAMG00000007807 | - | 97 | 42.490 | Mastacembelus_armatus |
| ENSG00000155363 | MOV10 | 95 | 45.126 | ENSMZEG00005027164 | - | 97 | 42.913 | Maylandia_zebra |
| ENSG00000155363 | MOV10 | 95 | 42.731 | ENSMZEG00005010177 | - | 96 | 42.731 | Maylandia_zebra |
| ENSG00000155363 | MOV10 | 94 | 49.779 | ENSMGAG00000004879 | MOV10 | 98 | 49.064 | Meleagris_gallopavo |
| ENSG00000155363 | MOV10 | 100 | 91.878 | ENSMAUG00000013906 | Mov10 | 100 | 91.534 | Mesocricetus_auratus |
| ENSG00000155363 | MOV10 | 100 | 96.116 | ENSMICG00000003187 | MOV10 | 100 | 96.116 | Microcebus_murinus |
| ENSG00000155363 | MOV10 | 100 | 92.194 | ENSMOCG00000007110 | Mov10 | 100 | 91.434 | Microtus_ochrogaster |
| ENSG00000155363 | MOV10 | 86 | 42.169 | ENSMMOG00000008592 | MOV10 | 91 | 48.730 | Mola_mola |
| ENSG00000155363 | MOV10 | 95 | 44.884 | ENSMMOG00000008456 | - | 90 | 49.611 | Mola_mola |
| ENSG00000155363 | MOV10 | 100 | 68.349 | ENSMODG00000001008 | MOV10 | 97 | 67.925 | Monodelphis_domestica |
| ENSG00000155363 | MOV10 | 95 | 44.774 | ENSMALG00000013875 | mov10b.1 | 92 | 44.774 | Monopterus_albus |
| ENSG00000155363 | MOV10 | 100 | 91.878 | MGP_CAROLIEiJ_G0025467 | Mov10 | 100 | 91.434 | Mus_caroli |
| ENSG00000155363 | MOV10 | 100 | 91.667 | ENSMUSG00000002227 | Mov10 | 100 | 91.235 | Mus_musculus |
| ENSG00000155363 | MOV10 | 100 | 91.667 | MGP_PahariEiJ_G0026912 | Mov10 | 100 | 91.235 | Mus_pahari |
| ENSG00000155363 | MOV10 | 100 | 91.667 | MGP_SPRETEiJ_G0026416 | Mov10 | 100 | 91.235 | Mus_spretus |
| ENSG00000155363 | MOV10 | 100 | 94.509 | ENSMPUG00000004985 | MOV10 | 98 | 94.417 | Mustela_putorius_furo |
| ENSG00000155363 | MOV10 | 95 | 85.635 | ENSMLUG00000009815 | MOV10 | 99 | 85.635 | Myotis_lucifugus |
| ENSG00000155363 | MOV10 | 100 | 92.405 | ENSNGAG00000021868 | Mov10 | 100 | 91.932 | Nannospalax_galili |
| ENSG00000155363 | MOV10 | 97 | 41.449 | ENSNBRG00000018612 | mov10b.2 | 97 | 41.449 | Neolamprologus_brichardi |
| ENSG00000155363 | MOV10 | 94 | 95.703 | ENSOPRG00000009237 | MOV10 | 94 | 95.703 | Ochotona_princeps |
| ENSG00000155363 | MOV10 | 100 | 87.962 | ENSODEG00000010401 | MOV10 | 95 | 87.537 | Octodon_degus |
| ENSG00000155363 | MOV10 | 100 | 44.491 | ENSONIG00000012274 | mov10b.1 | 97 | 44.444 | Oreochromis_niloticus |
| ENSG00000155363 | MOV10 | 100 | 94.915 | ENSOCUG00000000276 | MOV10 | 100 | 94.915 | Oryctolagus_cuniculus |
| ENSG00000155363 | MOV10 | 65 | 41.868 | ENSORLG00000023540 | - | 69 | 41.868 | Oryzias_latipes |
| ENSG00000155363 | MOV10 | 65 | 42.029 | ENSORLG00020011008 | - | 69 | 42.029 | Oryzias_latipes_hni |
| ENSG00000155363 | MOV10 | 100 | 96.304 | ENSOGAG00000014080 | MOV10 | 100 | 96.211 | Otolemur_garnettii |
| ENSG00000155363 | MOV10 | 100 | 92.080 | ENSOARG00000019733 | MOV10 | 98 | 91.725 | Ovis_aries |
| ENSG00000155363 | MOV10 | 100 | 100.000 | ENSPPAG00000031079 | MOV10 | 100 | 100.000 | Pan_paniscus |
| ENSG00000155363 | MOV10 | 100 | 96.515 | ENSPPRG00000007950 | MOV10 | 100 | 96.411 | Panthera_pardus |
| ENSG00000155363 | MOV10 | 100 | 96.515 | ENSPTIG00000016249 | MOV10 | 100 | 96.515 | Panthera_tigris_altaica |
| ENSG00000155363 | MOV10 | 100 | 100.000 | ENSPTRG00000001111 | MOV10 | 100 | 100.000 | Pan_troglodytes |
| ENSG00000155363 | MOV10 | 100 | 95.713 | ENSPANG00000011819 | MOV10 | 100 | 95.713 | Papio_anubis |
| ENSG00000155363 | MOV10 | 95 | 44.342 | ENSPKIG00000019999 | - | 96 | 44.342 | Paramormyrops_kingsleyae |
| ENSG00000155363 | MOV10 | 66 | 45.227 | ENSPMGG00000007310 | - | 85 | 45.383 | Periophthalmus_magnuspinnatus |
| ENSG00000155363 | MOV10 | 100 | 91.245 | ENSPEMG00000007910 | Mov10 | 100 | 90.837 | Peromyscus_maniculatus_bairdii |
| ENSG00000155363 | MOV10 | 100 | 65.546 | ENSPCIG00000002681 | MOV10 | 93 | 65.098 | Phascolarctos_cinereus |
| ENSG00000155363 | MOV10 | 99 | 44.434 | ENSPFOG00000000853 | - | 99 | 44.434 | Poecilia_formosa |
| ENSG00000155363 | MOV10 | 80 | 35.576 | ENSPFOG00000007579 | - | 99 | 35.576 | Poecilia_formosa |
| ENSG00000155363 | MOV10 | 95 | 42.935 | ENSPFOG00000013423 | mov10a | 97 | 40.472 | Poecilia_formosa |
| ENSG00000155363 | MOV10 | 95 | 43.921 | ENSPLAG00000013130 | mov10a | 84 | 43.921 | Poecilia_latipinna |
| ENSG00000155363 | MOV10 | 99 | 43.384 | ENSPLAG00000023722 | mov10b.2 | 99 | 43.384 | Poecilia_latipinna |
| ENSG00000155363 | MOV10 | 90 | 45.539 | ENSPMEG00000022500 | mov10a | 74 | 45.539 | Poecilia_mexicana |
| ENSG00000155363 | MOV10 | 100 | 44.323 | ENSPMEG00000017607 | mov10b.1 | 99 | 44.140 | Poecilia_mexicana |
| ENSG00000155363 | MOV10 | 99 | 43.798 | ENSPREG00000020106 | mov10b.1 | 99 | 43.798 | Poecilia_reticulata |
| ENSG00000155363 | MOV10 | 92 | 43.390 | ENSPREG00000011301 | mov10a | 93 | 43.390 | Poecilia_reticulata |
| ENSG00000155363 | MOV10 | 100 | 99.701 | ENSPPYG00000001020 | MOV10 | 100 | 99.701 | Pongo_abelii |
| ENSG00000155363 | MOV10 | 93 | 95.556 | ENSPCAG00000003786 | MOV10 | 93 | 95.556 | Procavia_capensis |
| ENSG00000155363 | MOV10 | 100 | 96.813 | ENSPCOG00000005180 | MOV10 | 100 | 96.813 | Propithecus_coquereli |
| ENSG00000155363 | MOV10 | 100 | 89.546 | ENSPVAG00000002477 | MOV10 | 100 | 88.933 | Pteropus_vampyrus |
| ENSG00000155363 | MOV10 | 95 | 45.016 | ENSPNYG00000016801 | mov10b.1 | 97 | 42.717 | Pundamilia_nyererei |
| ENSG00000155363 | MOV10 | 99 | 43.816 | ENSPNAG00000017939 | MOV10 | 99 | 43.816 | Pygocentrus_nattereri |
| ENSG00000155363 | MOV10 | 95 | 45.585 | ENSPNAG00000017916 | - | 97 | 43.393 | Pygocentrus_nattereri |
| ENSG00000155363 | MOV10 | 100 | 91.983 | ENSRNOG00000012946 | Mov10 | 100 | 91.534 | Rattus_norvegicus |
| ENSG00000155363 | MOV10 | 100 | 99.103 | ENSRBIG00000040802 | MOV10 | 100 | 99.103 | Rhinopithecus_bieti |
| ENSG00000155363 | MOV10 | 100 | 99.103 | ENSRROG00000041135 | MOV10 | 100 | 99.103 | Rhinopithecus_roxellana |
| ENSG00000155363 | MOV10 | 100 | 98.704 | ENSSBOG00000005229 | MOV10 | 100 | 98.704 | Saimiri_boliviensis_boliviensis |
| ENSG00000155363 | MOV10 | 100 | 67.684 | ENSSHAG00000000450 | MOV10 | 97 | 67.604 | Sarcophilus_harrisii |
| ENSG00000155363 | MOV10 | 97 | 47.854 | ENSSFOG00015008568 | mov10 | 99 | 45.525 | Scleropages_formosus |
| ENSG00000155363 | MOV10 | 65 | 47.191 | ENSSMAG00000004913 | - | 91 | 47.191 | Scophthalmus_maximus |
| ENSG00000155363 | MOV10 | 99 | 48.346 | ENSSPUG00000013262 | MOV10 | 99 | 48.346 | Sphenodon_punctatus |
| ENSG00000155363 | MOV10 | 100 | 93.664 | ENSSSCG00000006776 | MOV10 | 100 | 94.083 | Sus_scrofa |
| ENSG00000155363 | MOV10 | 94 | 48.201 | ENSTGUG00000017534 | MOV10 | 98 | 48.201 | Taeniopygia_guttata |
| ENSG00000155363 | MOV10 | 100 | 95.459 | ENSUAMG00000006720 | MOV10 | 100 | 95.314 | Ursus_americanus |
| ENSG00000155363 | MOV10 | 100 | 93.664 | ENSUMAG00000014721 | MOV10 | 100 | 94.675 | Ursus_maritimus |
| ENSG00000155363 | MOV10 | 100 | 94.931 | ENSVVUG00000016071 | MOV10 | 100 | 94.931 | Vulpes_vulpes |
| ENSG00000155363 | MOV10 | 99 | 43.556 | ENSXETG00000030428 | - | 99 | 43.100 | Xenopus_tropicalis |
| ENSG00000155363 | MOV10 | 99 | 50.052 | ENSXETG00000006901 | mov10 | 99 | 48.865 | Xenopus_tropicalis |
| ENSG00000155363 | MOV10 | 64 | 40.826 | ENSXCOG00000005350 | - | 84 | 40.826 | Xiphophorus_couchianus |
| ENSG00000155363 | MOV10 | 98 | 45.148 | ENSXMAG00000024782 | mov10b.1 | 99 | 43.466 | Xiphophorus_maculatus |
| ENSG00000155363 | MOV10 | 96 | 43.974 | ENSXMAG00000000875 | mov10a | 84 | 43.974 | Xiphophorus_maculatus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000932 | P-body | 16289642.24726324. | IDA | Component |
| GO:0003723 | RNA binding | 22658674.22681889. | HDA | Function |
| GO:0003723 | RNA binding | 21873635. | IBA | Function |
| GO:0003723 | RNA binding | 23093941.24726324. | IDA | Function |
| GO:0005515 | protein binding | 16289642.17507929.18552826.20543829.21282530.21903422.23093941.23125361.24726324.25464849.30122351. | IPI | Function |
| GO:0005524 | ATP binding | - | IEA | Function |
| GO:0005615 | extracellular space | 22664934. | HDA | Component |
| GO:0005634 | nucleus | - | ISS | Component |
| GO:0005829 | cytosol | 21873635. | IBA | Component |
| GO:0005829 | cytosol | - | IDA | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0007223 | Wnt signaling pathway, calcium modulating pathway | - | TAS | Process |
| GO:0010494 | cytoplasmic stress granule | - | IEA | Component |
| GO:0010526 | negative regulation of transposition, RNA-mediated | 23093941.30122351. | IDA | Process |
| GO:0010628 | positive regulation of gene expression | - | TAS | Process |
| GO:0010629 | negative regulation of gene expression | - | TAS | Process |
| GO:0032575 | ATP-dependent 5'-3' RNA helicase activity | 24726324. | IDA | Function |
| GO:0035194 | posttranscriptional gene silencing by RNA | 21873635. | IBA | Process |
| GO:0035195 | gene silencing by miRNA | 17507929. | IMP | Process |
| GO:0035279 | mRNA cleavage involved in gene silencing by miRNA | 16289642. | IDA | Process |
| GO:0036464 | cytoplasmic ribonucleoprotein granule | 30122351. | IDA | Component |
| GO:0043186 | P granule | 21873635. | IBA | Component |
| GO:0045652 | regulation of megakaryocyte differentiation | - | TAS | Process |
| GO:0061014 | positive regulation of mRNA catabolic process | 24726324. | IDA | Process |
| GO:0061158 | 3'-UTR-mediated mRNA destabilization | 24726324. | IDA | Process |
| GO:0150011 | regulation of neuron projection arborization | - | ISS | Process |