
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000558814 | MYO1E-205 | 768 | - | - | - (aa) | - | - |
| ENST00000288235 | MYO1E-201 | 8673 | - | ENSP00000288235 | 1108 (aa) | - | Q12965 |
| ENST00000559412 | MYO1E-207 | 541 | - | ENSP00000453936 | 73 (aa) | - | H0YNB0 |
| ENST00000559269 | MYO1E-206 | 828 | - | ENSP00000453232 | 276 (aa) | - | H0YLJ4 |
| ENST00000558571 | MYO1E-203 | 557 | - | ENSP00000453811 | 112 (aa) | - | H0YN00 |
| ENST00000560642 | MYO1E-209 | 559 | - | - | - (aa) | - | - |
| ENST00000558646 | MYO1E-204 | 578 | - | - | - (aa) | - | - |
| ENST00000558182 | MYO1E-202 | 544 | - | - | - (aa) | - | - |
| ENST00000559489 | MYO1E-208 | 562 | - | ENSP00000453178 | 187 (aa) | - | H0YLE5 |
| ENST00000560749 | MYO1E-210 | 952 | - | ENSP00000454113 | 317 (aa) | - | H0YNQ8 |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000157483 | Alcoholism | 5.73905e-005 | 22978509 |
| ENSG00000157483 | Alcoholism | 1.26726e-005 | 22978509 |
| ENSG00000157483 | Blood Pressure | 9.0620000E-006 | - |
| ENSG00000157483 | Blood Pressure | 1.6510000E-005 | - |
| ENSG00000157483 | Blood Pressure | 7.5330000E-005 | - |
| ENSG00000157483 | Platelet Function Tests | 2.3700000E-010 | - |
| ENSG00000157483 | Platelet Function Tests | 8.5600000E-006 | - |
| ENSG00000157483 | Platelet Function Tests | 6.8400000E-006 | - |
| ENSG00000157483 | Vascular Calcification | 1E-10 | 22916037 |
| ENSG00000157483 | Communication Disorders | 1E-7 | 24047820 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000157483 | rs2306786 | 15 | 59195731 | ? | Metabolite levels | 22916037 | EFO_0004723 | ||
| ENSG00000157483 | rs4218 | 15 | 59136459 | G | Social communication problems | 24047820 | [0.063-0.137] unit increase | 0.1 | EFO_0005427 |
| ENSG00000157483 | rs114625006 | 15 | 59361839 | ? | Coronary artery calcified atherosclerotic plaque (90 or 130 HU threshold) in type 2 diabetes | 29221444 | [0.085-0.309] unit increase | 0.197 | EFO_0004723|EFO_0001360 |
| ENSG00000157483 | rs78595738 | 15 | 59179319 | ? | Measles | 28928442 | [0.4-0.99] unit decrease | 0.6929 | EFO_0008414 |
| ENSG00000157483 | rs7175642 | 15 | 59157880 | ? | Body mass index | 30595370 | EFO_0004340 | ||
| ENSG00000157483 | rs3191402 | 15 | 59136961 | A | Pulse pressure | 30224653 | [0.084-0.155] unit decrease | 0.1191 | EFO_0005763 |
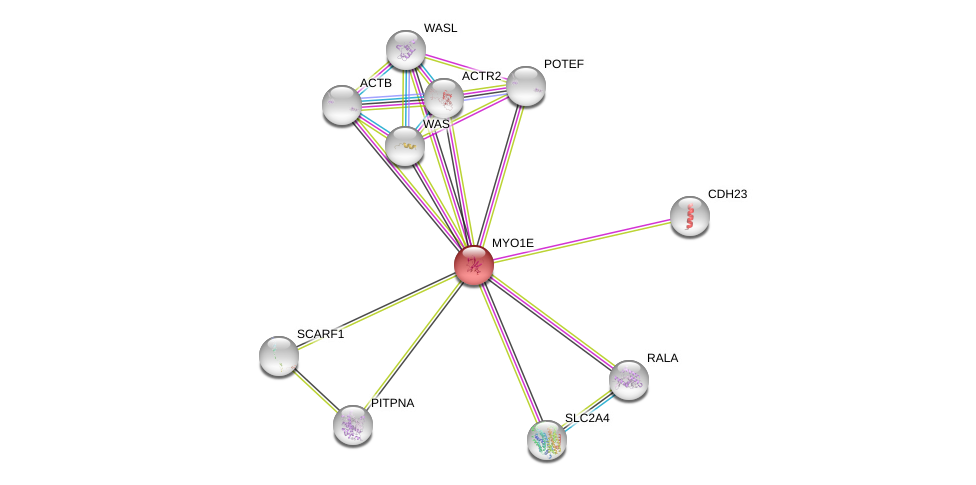
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000146 | microfilament motor activity | 7932763. | TAS | Function |
| GO:0001570 | vasculogenesis | - | IEA | Process |
| GO:0001701 | in utero embryonic development | - | IEA | Process |
| GO:0003094 | glomerular filtration | - | ISS | Process |
| GO:0003774 | motor activity | 7932763. | TAS | Function |
| GO:0005515 | protein binding | 17257598.19846667.21458045. | IPI | Function |
| GO:0005516 | calmodulin binding | 11940582. | IDA | Function |
| GO:0005524 | ATP binding | - | IEA | Function |
| GO:0005737 | cytoplasm | - | ISS | Component |
| GO:0005856 | cytoskeleton | 17257598. | IDA | Component |
| GO:0005903 | brush border | - | IEA | Component |
| GO:0005911 | cell-cell junction | - | ISS | Component |
| GO:0005912 | adherens junction | - | ISS | Component |
| GO:0006807 | nitrogen compound metabolic process | - | IEA | Process |
| GO:0006897 | endocytosis | 17257598. | IMP | Process |
| GO:0015629 | actin cytoskeleton | 7932763. | TAS | Component |
| GO:0016459 | myosin complex | 11940582. | TAS | Component |
| GO:0030048 | actin filament-based movement | 7932763. | TAS | Process |
| GO:0032836 | glomerular basement membrane development | - | ISS | Process |
| GO:0035091 | phosphatidylinositol binding | 20860408. | IDA | Function |
| GO:0035166 | post-embryonic hemopoiesis | - | IEA | Process |
| GO:0042623 | ATPase activity, coupled | 11940582. | IDA | Function |
| GO:0045334 | clathrin-coated endocytic vesicle | 17257598. | IDA | Component |
| GO:0048008 | platelet-derived growth factor receptor signaling pathway | - | IEA | Process |
| GO:0051015 | actin filament binding | 11940582. | IDA | Function |
| GO:0070062 | extracellular exosome | 20458337. | HDA | Component |
| GO:0072015 | glomerular visceral epithelial cell development | - | ISS | Process |