
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000497281 | G6PD-210 | 532 | - | - | - (aa) | - | - |
| ENST00000488434 | G6PD-207 | 432 | - | - | - (aa) | - | - |
| ENST00000433845 | G6PD-204 | 817 | - | ENSP00000394690 | 256 (aa) | - | E9PD92 |
| ENST00000393564 | G6PD-203 | 1661 | - | ENSP00000377194 | 515 (aa) | - | P11413 |
| ENST00000393562 | G6PD-202 | 2223 | - | ENSP00000377192 | 515 (aa) | - | P11413 |
| ENST00000647501 | G6PD-212 | 471 | - | - | - (aa) | - | - |
| ENST00000439227 | G6PD-205 | 1074 | - | ENSP00000395599 | 338 (aa) | - | E7EUI8 |
| ENST00000621232 | G6PD-211 | 2632 | - | ENSP00000483686 | 515 (aa) | - | P11413 |
| ENST00000440967 | G6PD-206 | 1056 | - | ENSP00000400648 | 320 (aa) | - | E7EM57 |
| ENST00000489497 | G6PD-208 | 512 | - | - | - (aa) | - | - |
| ENST00000490651 | G6PD-209 | 783 | - | - | - (aa) | - | - |
| ENST00000369620 | G6PD-201 | 1799 | - | ENSP00000358633 | 561 (aa) | - | P11413 |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000160211 | Erythrocyte Count | 4E-13 | 23696099 |
| ENSG00000160211 | Erythrocyte Count | 1E-14 | 23696099 |
| ENSG00000160211 | Erythrocyte Indices | 9E-9 | 23696099 |
| ENSG00000160211 | Hematocrit | 2E-16 | 23446634 |
| ENSG00000160211 | Hemoglobins | 4E-18 | 23446634 |
| ENSG00000160211 | Erythrocyte Count | 4E-14 | 23446634 |
| ENSG00000160211 | Hemoglobin A, Glycosylated | 3E-11 | 26366553 |
| ENSG00000160211 | Hemoglobins | 3E-11 | 26366553 |
| ENSG00000160211 | Erythrocyte Count | 2E-11 | 23446634 |
| ENSG00000160211 | Erythrocyte Count | 4E-19 | 23446634 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000160211 | rs762516 | X | 154536448 | T | Red blood cell traits | 23446634 | [0.34-0.56] % decrease | 0.452 | EFO_0004348 |
| ENSG00000160211 | rs762516 | X | 154536448 | T | Red blood cell traits | 23446634 | [0.12-0.2] g/dL decrease | 0.1614 | EFO_0004509 |
| ENSG00000160211 | rs762516 | X | 154536448 | T | Red blood cell traits | 23446634 | [1.17-1.99] fL increase | 1.5768 | EFO_0004526 |
| ENSG00000160211 | rs1050828 | X | 154536002 | T | Red blood cell traits | 23446634 | [0.023-0.042] % decrease | 0.0326 | EFO_0005192 |
| ENSG00000160211 | rs5030868 | X | 154534419 | A | Hemoglobin levels | 26366553 | [0.088-0.163] g/dl decrease | 0.1256 | EFO_0004509|EFO_0007629 |
| ENSG00000160211 | rs1050828 | X | 154536002 | A | Red blood cell traits | 23696099 | [0.14-0.26] x10^12/L decrease | 0.2 | EFO_0004305 |
| ENSG00000160211 | rs1050828 | X | 154536002 | A | Red blood cell traits | 23696099 | [1.83-3.09] fL increase | 2.46 | EFO_0004526 |
| ENSG00000160211 | rs1050828 | X | 154536002 | A | Red blood cell traits | 23696099 | [0.48-0.96] pg increase | 0.72 | EFO_0004527 |
| ENSG00000160211 | rs1050828 | X | 154536002 | T | Glycated hemoglobin levels | 28898252 | [0.35-0.41] unit decrease | 0.3819 | EFO_0004541 |
| ENSG00000160211 | rs1050828 | X | 154536002 | C | Iron status biomarkers (transferrin saturation) | 28334935 | [0.13-0.3] unit decrease | 0.21441 | EFO_0006333 |
| ENSG00000160211 | rs1050828 | X | 154536002 | C | Red blood cell count | 28453575 | [0.11-0.15] cells x E9 increase | 0.13 | EFO_0004305 |
| ENSG00000160211 | rs1050828 | X | 154536002 | C | Mean corpuscular volume | 28453575 | [1.49-2.35] fL decrease | 1.92 | EFO_0004526 |
| ENSG00000160211 | rs1050828 | X | 154536002 | A | Mean corpuscular hemoglobin | 28453575 | [0.38-0.71] pg increase | 0.546 | EFO_0004527 |
| ENSG00000160211 | rs1050828 | X | 154536002 | T | Red cell distribution width | 28453575 | [0.031-0.043] unit decrease | 0.04 | EFO_0005192 |
| ENSG00000160211 | rs1050828 | X | 154536002 | C | Mean corpuscular volume | 28453575 | [1.49-2.35] fL decrease | 1.92 | EFO_0004526 |
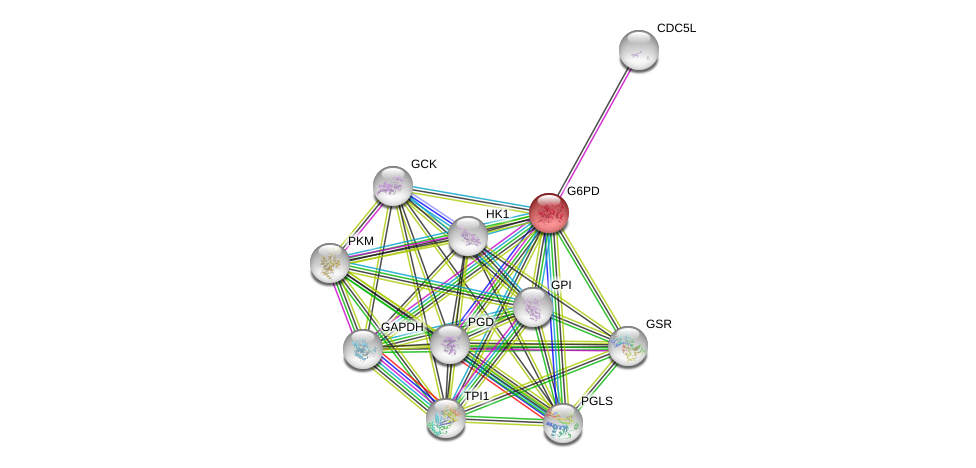
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000160211 | G6PD | 93 | 60.986 | WBGene00007108 | gspd-1 | 93 | 60.986 | Caenorhabditis_elegans |
| ENSG00000160211 | G6PD | 94 | 47.853 | YNL241C | ZWF1 | 94 | 47.853 | Saccharomyces_cerevisiae |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0004345 | glucose-6-phosphate dehydrogenase activity | 15858258. | IDA | Function |
| GO:0004345 | glucose-6-phosphate dehydrogenase activity | 743300.2420826.5643703. | IMP | Function |
| GO:0005515 | protein binding | 21157431.24769394. | IPI | Function |
| GO:0005536 | glucose binding | 15858258. | IDA | Function |
| GO:0005536 | glucose binding | 5643703. | IMP | Function |
| GO:0005634 | nucleus | - | IEA | Component |
| GO:0005737 | cytoplasm | - | IDA | Component |
| GO:0005829 | cytosol | 743300. | IDA | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0006006 | glucose metabolic process | - | IEA | Process |
| GO:0006098 | pentose-phosphate shunt | 2297768. | IDA | Process |
| GO:0006098 | pentose-phosphate shunt | - | TAS | Process |
| GO:0006629 | lipid metabolic process | 17361089. | TAS | Process |
| GO:0006695 | cholesterol biosynthetic process | 12027950. | IMP | Process |
| GO:0006739 | NADP metabolic process | 15858258. | IDA | Process |
| GO:0006740 | NADPH regeneration | 17516514. | IMP | Process |
| GO:0006749 | glutathione metabolic process | 2420826.17516514. | IMP | Process |
| GO:0009051 | pentose-phosphate shunt, oxidative branch | 2420826. | IMP | Process |
| GO:0009898 | cytoplasmic side of plasma membrane | 743300. | IDA | Component |
| GO:0010041 | response to iron(III) ion | - | IEA | Process |
| GO:0010734 | negative regulation of protein glutathionylation | 17516514. | IMP | Process |
| GO:0014070 | response to organic cyclic compound | - | IEA | Process |
| GO:0016020 | membrane | 19946888. | HDA | Component |
| GO:0019322 | pentose biosynthetic process | 5643703. | IDA | Process |
| GO:0021762 | substantia nigra development | 22926577. | HEP | Process |
| GO:0032094 | response to food | - | IEA | Process |
| GO:0034599 | cellular response to oxidative stress | 17516514. | IMP | Process |
| GO:0042802 | identical protein binding | 24769394.25416956. | IPI | Function |
| GO:0042803 | protein homodimerization activity | 15858258. | IPI | Function |
| GO:0043249 | erythrocyte maturation | 5643703. | IMP | Process |
| GO:0043523 | regulation of neuron apoptotic process | - | IEA | Process |
| GO:0045471 | response to ethanol | - | IEA | Process |
| GO:0046390 | ribose phosphate biosynthetic process | 2420826. | IMP | Process |
| GO:0050661 | NADP binding | 15858258. | IDA | Function |
| GO:0051156 | glucose 6-phosphate metabolic process | 15858258. | IDA | Process |
| GO:0051156 | glucose 6-phosphate metabolic process | 5643703. | IMP | Process |
| GO:0055114 | oxidation-reduction process | 2420826. | IMP | Process |
| GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development | - | IEA | Process |
| GO:0070062 | extracellular exosome | 19056867.23533145. | HDA | Component |
| GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel | - | IEA | Process |
| GO:2000378 | negative regulation of reactive oxygen species metabolic process | - | IEA | Process |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| ACC | |||||||
| ACC | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| LAML | |||||||
| LAML | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| MESO | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| CESC | |||
| DLBC | |||
| HNSC | |||
| KIRC | |||
| LIHC | |||
| LUAD | |||
| LUSC | |||
| OV | |||
| PAAD | |||
| READ | |||
| STAD | |||
| TGCT | |||
| THCA | |||
| UCS |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| MESO | ||
| ACC | ||
| HNSC | ||
| BRCA | ||
| KIRP | ||
| BLCA | ||
| CESC | ||
| LAML | ||
| LIHC |