
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000295920 | tRNA_Me_trans | PF03054.16 | 2.6e-05 | 1 | 1 |
| ENSP00000419851 | tRNA_Me_trans | PF03054.16 | 3.4e-05 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000295920 | GMPS-201 | 2024 | - | ENSP00000295920 | 594 (aa) | - | P49915 |
| ENST00000476145 | GMPS-202 | 570 | - | - | - (aa) | - | - |
| ENST00000496455 | GMPS-203 | 8751 | XM_011513263 | ENSP00000419851 | 693 (aa) | XP_011511565 | P49915 |
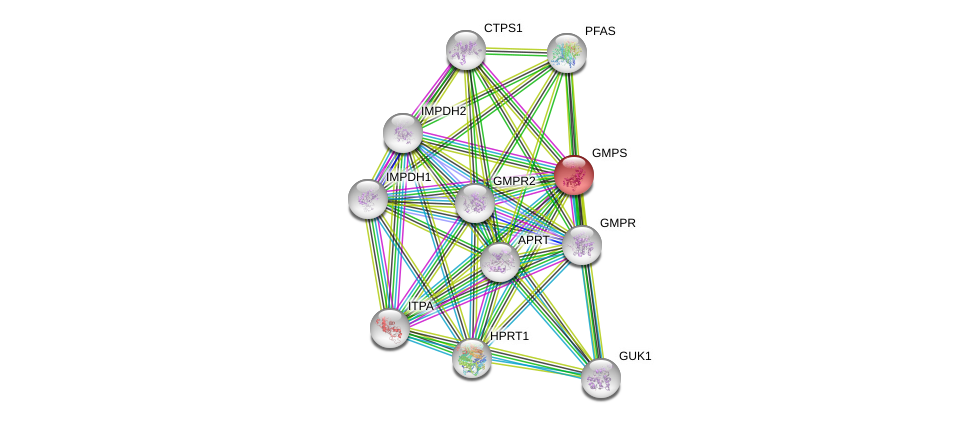
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000163655 | GMPS | 100 | 86.003 | ENSAPOG00000010728 | gmps | 100 | 86.003 | Acanthochromis_polyacanthus |
| ENSG00000163655 | GMPS | 100 | 99.134 | ENSAMEG00000002551 | GMPS | 100 | 99.134 | Ailuropoda_melanoleuca |
| ENSG00000163655 | GMPS | 100 | 85.185 | ENSACIG00000012452 | gmps | 97 | 87.145 | Amphilophus_citrinellus |
| ENSG00000163655 | GMPS | 100 | 86.436 | ENSAOCG00000003292 | gmps | 100 | 86.436 | Amphiprion_ocellaris |
| ENSG00000163655 | GMPS | 100 | 86.147 | ENSAPEG00000020189 | gmps | 100 | 86.147 | Amphiprion_percula |
| ENSG00000163655 | GMPS | 100 | 86.195 | ENSATEG00000002414 | gmps | 99 | 86.082 | Anabas_testudineus |
| ENSG00000163655 | GMPS | 99 | 94.737 | ENSACAG00000000556 | GMPS | 99 | 94.737 | Anolis_carolinensis |
| ENSG00000163655 | GMPS | 100 | 99.278 | ENSANAG00000036394 | GMPS | 100 | 99.278 | Aotus_nancymaae |
| ENSG00000163655 | GMPS | 100 | 86.291 | ENSACLG00000003086 | gmps | 100 | 86.291 | Astatotilapia_calliptera |
| ENSG00000163655 | GMPS | 100 | 86.580 | ENSAMXG00000029474 | gmps | 100 | 86.580 | Astyanax_mexicanus |
| ENSG00000163655 | GMPS | 100 | 98.846 | ENSBTAG00000013013 | GMPS | 100 | 98.846 | Bos_taurus |
| ENSG00000163655 | GMPS | 100 | 48.160 | WBGene00010912 | gmps-1 | 97 | 51.003 | Caenorhabditis_elegans |
| ENSG00000163655 | GMPS | 100 | 99.134 | ENSCJAG00000000863 | GMPS | 100 | 99.134 | Callithrix_jacchus |
| ENSG00000163655 | GMPS | 100 | 97.315 | ENSCAFG00000008780 | GMPS | 97 | 98.318 | Canis_familiaris |
| ENSG00000163655 | GMPS | 100 | 98.701 | ENSCAFG00020005568 | GMPS | 100 | 98.701 | Canis_lupus_dingo |
| ENSG00000163655 | GMPS | 100 | 98.557 | ENSCHIG00000022844 | GMPS | 100 | 98.557 | Capra_hircus |
| ENSG00000163655 | GMPS | 100 | 97.475 | ENSTSYG00000009457 | GMPS | 97 | 98.246 | Carlito_syrichta |
| ENSG00000163655 | GMPS | 90 | 93.691 | ENSCAPG00000008634 | GMPS | 85 | 93.691 | Cavia_aperea |
| ENSG00000163655 | GMPS | 97 | 96.562 | ENSCPOG00000011574 | GMPS | 98 | 96.562 | Cavia_porcellus |
| ENSG00000163655 | GMPS | 100 | 99.158 | ENSCCAG00000031267 | GMPS | 100 | 99.158 | Cebus_capucinus |
| ENSG00000163655 | GMPS | 100 | 99.856 | ENSCATG00000031121 | GMPS | 100 | 99.856 | Cercocebus_atys |
| ENSG00000163655 | GMPS | 100 | 97.138 | ENSCLAG00000007499 | GMPS | 100 | 97.661 | Chinchilla_lanigera |
| ENSG00000163655 | GMPS | 100 | 99.856 | ENSCSAG00000012022 | GMPS | 100 | 99.856 | Chlorocebus_sabaeus |
| ENSG00000163655 | GMPS | 99 | 75.585 | ENSCHOG00000008585 | GMPS | 100 | 75.585 | Choloepus_hoffmanni |
| ENSG00000163655 | GMPS | 100 | 93.795 | ENSCPBG00000018525 | GMPS | 100 | 93.795 | Chrysemys_picta_bellii |
| ENSG00000163655 | GMPS | 99 | 88.450 | ENSCANG00000027748 | GMPS | 97 | 98.280 | Colobus_angolensis_palliatus |
| ENSG00000163655 | GMPS | 100 | 97.547 | ENSCGRG00001018772 | Gmps | 100 | 97.547 | Cricetulus_griseus_chok1gshd |
| ENSG00000163655 | GMPS | 100 | 97.547 | ENSCGRG00000002986 | Gmps | 100 | 97.547 | Cricetulus_griseus_crigri |
| ENSG00000163655 | GMPS | 100 | 85.017 | ENSCSEG00000001844 | gmps | 100 | 84.704 | Cynoglossus_semilaevis |
| ENSG00000163655 | GMPS | 100 | 86.364 | ENSCVAG00000013802 | gmps | 100 | 86.291 | Cyprinodon_variegatus |
| ENSG00000163655 | GMPS | 100 | 86.869 | ENSDARG00000079848 | gmps | 100 | 86.869 | Danio_rerio |
| ENSG00000163655 | GMPS | 100 | 98.990 | ENSDNOG00000017772 | GMPS | 100 | 98.990 | Dasypus_novemcinctus |
| ENSG00000163655 | GMPS | 100 | 98.124 | ENSDORG00000014996 | Gmps | 100 | 98.124 | Dipodomys_ordii |
| ENSG00000163655 | GMPS | 86 | 97.456 | ENSETEG00000010604 | - | 100 | 97.456 | Echinops_telfairi |
| ENSG00000163655 | GMPS | 100 | 97.691 | ENSEASG00005022509 | GMPS | 100 | 97.691 | Equus_asinus_asinus |
| ENSG00000163655 | GMPS | 100 | 97.980 | ENSECAG00000023269 | GMPS | 100 | 97.980 | Equus_caballus |
| ENSG00000163655 | GMPS | 99 | 97.515 | ENSEEUG00000011718 | GMPS | 100 | 97.515 | Erinaceus_europaeus |
| ENSG00000163655 | GMPS | 100 | 86.291 | ENSELUG00000008772 | gmps | 100 | 86.291 | Esox_lucius |
| ENSG00000163655 | GMPS | 100 | 99.158 | ENSFCAG00000025327 | GMPS | 100 | 99.158 | Felis_catus |
| ENSG00000163655 | GMPS | 99 | 93.704 | ENSFALG00000004855 | GMPS | 99 | 93.704 | Ficedula_albicollis |
| ENSG00000163655 | GMPS | 100 | 96.970 | ENSFDAG00000009186 | GMPS | 99 | 97.106 | Fukomys_damarensis |
| ENSG00000163655 | GMPS | 100 | 85.714 | ENSFHEG00000023029 | gmps | 100 | 85.714 | Fundulus_heteroclitus |
| ENSG00000163655 | GMPS | 100 | 83.670 | ENSGMOG00000009029 | gmps | 98 | 85.650 | Gadus_morhua |
| ENSG00000163655 | GMPS | 100 | 94.661 | ENSGALG00000010284 | GMPS | 91 | 94.661 | Gallus_gallus |
| ENSG00000163655 | GMPS | 100 | 83.333 | ENSGACG00000007053 | gmps | 99 | 81.402 | Gasterosteus_aculeatus |
| ENSG00000163655 | GMPS | 100 | 93.506 | ENSGAGG00000019485 | GMPS | 100 | 93.506 | Gopherus_agassizii |
| ENSG00000163655 | GMPS | 100 | 99.856 | ENSGGOG00000017082 | GMPS | 100 | 99.856 | Gorilla_gorilla |
| ENSG00000163655 | GMPS | 100 | 86.436 | ENSHBUG00000000946 | gmps | 100 | 86.436 | Haplochromis_burtoni |
| ENSG00000163655 | GMPS | 100 | 98.124 | ENSHGLG00000019482 | GMPS | 100 | 98.124 | Heterocephalus_glaber_female |
| ENSG00000163655 | GMPS | 100 | 97.306 | ENSHGLG00100019096 | - | 100 | 98.099 | Heterocephalus_glaber_male |
| ENSG00000163655 | GMPS | 100 | 85.354 | ENSHCOG00000011768 | gmps | 100 | 85.137 | Hippocampus_comes |
| ENSG00000163655 | GMPS | 100 | 82.155 | ENSIPUG00000015059 | gmps | 99 | 83.043 | Ictalurus_punctatus |
| ENSG00000163655 | GMPS | 99 | 98.392 | ENSSTOG00000026541 | GMPS | 100 | 98.392 | Ictidomys_tridecemlineatus |
| ENSG00000163655 | GMPS | 100 | 98.701 | ENSJJAG00000023127 | Gmps | 100 | 98.701 | Jaculus_jaculus |
| ENSG00000163655 | GMPS | 100 | 86.003 | ENSKMAG00000020846 | gmps | 100 | 86.003 | Kryptolebias_marmoratus |
| ENSG00000163655 | GMPS | 100 | 86.027 | ENSLBEG00000011676 | gmps | 100 | 86.003 | Labrus_bergylta |
| ENSG00000163655 | GMPS | 100 | 90.572 | ENSLACG00000007529 | GMPS | 99 | 91.336 | Latimeria_chalumnae |
| ENSG00000163655 | GMPS | 100 | 88.456 | ENSLOCG00000003174 | gmps | 100 | 88.456 | Lepisosteus_oculatus |
| ENSG00000163655 | GMPS | 100 | 97.114 | ENSLAFG00000012230 | GMPS | 100 | 97.114 | Loxodonta_africana |
| ENSG00000163655 | GMPS | 100 | 99.832 | ENSMFAG00000039350 | GMPS | 100 | 99.832 | Macaca_fascicularis |
| ENSG00000163655 | GMPS | 100 | 99.832 | ENSMMUG00000007324 | GMPS | 100 | 99.832 | Macaca_mulatta |
| ENSG00000163655 | GMPS | 99 | 88.081 | ENSMNEG00000029760 | GMPS | 100 | 88.081 | Macaca_nemestrina |
| ENSG00000163655 | GMPS | 100 | 86.580 | ENSMAMG00000003973 | gmps | 100 | 86.580 | Mastacembelus_armatus |
| ENSG00000163655 | GMPS | 100 | 86.291 | ENSMZEG00005012210 | gmps | 100 | 86.291 | Maylandia_zebra |
| ENSG00000163655 | GMPS | 100 | 93.603 | ENSMGAG00000010733 | GMPS | 98 | 94.075 | Meleagris_gallopavo |
| ENSG00000163655 | GMPS | 100 | 98.822 | ENSMICG00000049568 | GMPS | 100 | 98.822 | Microcebus_murinus |
| ENSG00000163655 | GMPS | 100 | 97.403 | ENSMOCG00000021456 | Gmps | 100 | 97.403 | Microtus_ochrogaster |
| ENSG00000163655 | GMPS | 100 | 84.680 | ENSMMOG00000004360 | gmps | 100 | 84.416 | Mola_mola |
| ENSG00000163655 | GMPS | 99 | 96.204 | ENSMODG00000015770 | GMPS | 99 | 96.204 | Monodelphis_domestica |
| ENSG00000163655 | GMPS | 100 | 86.147 | ENSMALG00000009440 | gmps | 100 | 86.147 | Monopterus_albus |
| ENSG00000163655 | GMPS | 100 | 98.124 | MGP_CAROLIEiJ_G0025038 | Gmps | 100 | 98.124 | Mus_caroli |
| ENSG00000163655 | GMPS | 100 | 97.980 | ENSMUSG00000027823 | Gmps | 100 | 97.980 | Mus_musculus |
| ENSG00000163655 | GMPS | 100 | 97.691 | MGP_PahariEiJ_G0026482 | Gmps | 100 | 97.691 | Mus_pahari |
| ENSG00000163655 | GMPS | 75 | 97.885 | MGP_SPRETEiJ_G0025978 | - | 98 | 97.885 | Mus_spretus |
| ENSG00000163655 | GMPS | 100 | 98.846 | ENSMPUG00000002781 | GMPS | 100 | 98.846 | Mustela_putorius_furo |
| ENSG00000163655 | GMPS | 100 | 96.296 | ENSMLUG00000015442 | GMPS | 100 | 96.248 | Myotis_lucifugus |
| ENSG00000163655 | GMPS | 100 | 97.403 | ENSNGAG00000019127 | Gmps | 100 | 97.403 | Nannospalax_galili |
| ENSG00000163655 | GMPS | 100 | 86.291 | ENSNBRG00000001913 | gmps | 100 | 86.291 | Neolamprologus_brichardi |
| ENSG00000163655 | GMPS | 100 | 99.832 | ENSNLEG00000003856 | GMPS | 100 | 99.832 | Nomascus_leucogenys |
| ENSG00000163655 | GMPS | 100 | 96.465 | ENSODEG00000005062 | GMPS | 100 | 97.432 | Octodon_degus |
| ENSG00000163655 | GMPS | 100 | 86.436 | ENSONIG00000005518 | gmps | 100 | 86.436 | Oreochromis_niloticus |
| ENSG00000163655 | GMPS | 99 | 96.075 | ENSOANG00000013670 | GMPS | 100 | 96.252 | Ornithorhynchus_anatinus |
| ENSG00000163655 | GMPS | 100 | 98.701 | ENSOCUG00000001480 | GMPS | 100 | 98.701 | Oryctolagus_cuniculus |
| ENSG00000163655 | GMPS | 100 | 84.512 | ENSORLG00000012593 | gmps | 100 | 84.127 | Oryzias_latipes |
| ENSG00000163655 | GMPS | 100 | 84.175 | ENSORLG00020012393 | gmps | 100 | 83.838 | Oryzias_latipes_hni |
| ENSG00000163655 | GMPS | 100 | 84.512 | ENSORLG00015020602 | gmps | 100 | 84.127 | Oryzias_latipes_hsok |
| ENSG00000163655 | GMPS | 100 | 85.690 | ENSOMEG00000004911 | gmps | 100 | 85.281 | Oryzias_melastigma |
| ENSG00000163655 | GMPS | 99 | 96.459 | ENSOGAG00000008523 | GMPS | 99 | 96.959 | Otolemur_garnettii |
| ENSG00000163655 | GMPS | 100 | 97.691 | ENSOARG00000003146 | GMPS | 100 | 97.691 | Ovis_aries |
| ENSG00000163655 | GMPS | 100 | 99.711 | ENSPPAG00000042787 | GMPS | 100 | 99.711 | Pan_paniscus |
| ENSG00000163655 | GMPS | 100 | 99.158 | ENSPPRG00000024889 | GMPS | 100 | 99.158 | Panthera_pardus |
| ENSG00000163655 | GMPS | 99 | 97.810 | ENSPTIG00000012623 | GMPS | 100 | 97.810 | Panthera_tigris_altaica |
| ENSG00000163655 | GMPS | 100 | 99.856 | ENSPANG00000017625 | GMPS | 100 | 99.856 | Papio_anubis |
| ENSG00000163655 | GMPS | 100 | 84.271 | ENSPKIG00000024697 | gmps | 100 | 84.271 | Paramormyrops_kingsleyae |
| ENSG00000163655 | GMPS | 100 | 93.651 | ENSPSIG00000007196 | GMPS | 100 | 93.651 | Pelodiscus_sinensis |
| ENSG00000163655 | GMPS | 100 | 85.690 | ENSPMGG00000015771 | gmps | 100 | 85.570 | Periophthalmus_magnuspinnatus |
| ENSG00000163655 | GMPS | 99 | 97.657 | ENSPEMG00000009799 | Gmps | 99 | 97.657 | Peromyscus_maniculatus_bairdii |
| ENSG00000163655 | GMPS | 100 | 96.825 | ENSPCIG00000000036 | GMPS | 100 | 96.825 | Phascolarctos_cinereus |
| ENSG00000163655 | GMPS | 100 | 85.354 | ENSPLAG00000015928 | gmps | 100 | 85.137 | Poecilia_latipinna |
| ENSG00000163655 | GMPS | 100 | 85.714 | ENSPREG00000016715 | gmps | 100 | 85.714 | Poecilia_reticulata |
| ENSG00000163655 | GMPS | 100 | 90.017 | ENSPPYG00000014229 | GMPS | 100 | 91.587 | Pongo_abelii |
| ENSG00000163655 | GMPS | 100 | 98.822 | ENSPCOG00000024460 | GMPS | 100 | 98.822 | Propithecus_coquereli |
| ENSG00000163655 | GMPS | 100 | 92.641 | ENSPVAG00000017458 | GMPS | 88 | 92.641 | Pteropus_vampyrus |
| ENSG00000163655 | GMPS | 100 | 86.291 | ENSPNYG00000010788 | gmps | 100 | 86.291 | Pundamilia_nyererei |
| ENSG00000163655 | GMPS | 100 | 86.291 | ENSPNAG00000014256 | gmps | 100 | 86.291 | Pygocentrus_nattereri |
| ENSG00000163655 | GMPS | 100 | 94.000 | ENSRNOG00000051151 | Gmps | 100 | 97.722 | Rattus_norvegicus |
| ENSG00000163655 | GMPS | 100 | 99.832 | ENSRBIG00000019515 | GMPS | 100 | 99.832 | Rhinopithecus_bieti |
| ENSG00000163655 | GMPS | 100 | 99.856 | ENSRROG00000035915 | GMPS | 100 | 99.856 | Rhinopithecus_roxellana |
| ENSG00000163655 | GMPS | 100 | 97.980 | ENSSBOG00000033046 | GMPS | 100 | 97.698 | Saimiri_boliviensis_boliviensis |
| ENSG00000163655 | GMPS | 100 | 95.118 | ENSSHAG00000010345 | GMPS | 99 | 96.093 | Sarcophilus_harrisii |
| ENSG00000163655 | GMPS | 100 | 86.580 | ENSSFOG00015001520 | gmps | 100 | 86.580 | Scleropages_formosus |
| ENSG00000163655 | GMPS | 100 | 85.690 | ENSSMAG00000020489 | gmps | 97 | 87.145 | Scophthalmus_maximus |
| ENSG00000163655 | GMPS | 100 | 85.690 | ENSSDUG00000011961 | gmps | 95 | 87.294 | Seriola_dumerili |
| ENSG00000163655 | GMPS | 100 | 86.003 | ENSSLDG00000017360 | gmps | 100 | 86.003 | Seriola_lalandi_dorsalis |
| ENSG00000163655 | GMPS | 100 | 94.661 | ENSSPUG00000015523 | - | 100 | 94.661 | Sphenodon_punctatus |
| ENSG00000163655 | GMPS | 100 | 86.291 | ENSSPAG00000017294 | gmps | 100 | 86.291 | Stegastes_partitus |
| ENSG00000163655 | GMPS | 100 | 99.278 | ENSSSCG00000028855 | GMPS | 100 | 99.278 | Sus_scrofa |
| ENSG00000163655 | GMPS | 100 | 93.651 | ENSTGUG00000011256 | GMPS | 100 | 93.651 | Taeniopygia_guttata |
| ENSG00000163655 | GMPS | 100 | 86.147 | ENSTRUG00000006141 | gmps | 100 | 86.147 | Takifugu_rubripes |
| ENSG00000163655 | GMPS | 100 | 85.092 | ENSTNIG00000003339 | gmps | 100 | 84.914 | Tetraodon_nigroviridis |
| ENSG00000163655 | GMPS | 99 | 90.936 | ENSTBEG00000000630 | GMPS | 100 | 90.936 | Tupaia_belangeri |
| ENSG00000163655 | GMPS | 100 | 99.134 | ENSUAMG00000015465 | GMPS | 100 | 99.134 | Ursus_americanus |
| ENSG00000163655 | GMPS | 99 | 97.515 | ENSUMAG00000011640 | GMPS | 99 | 97.515 | Ursus_maritimus |
| ENSG00000163655 | GMPS | 100 | 85.522 | ENSVPAG00000008478 | GMPS | 100 | 84.211 | Vicugna_pacos |
| ENSG00000163655 | GMPS | 100 | 97.811 | ENSVVUG00000019829 | GMPS | 99 | 98.113 | Vulpes_vulpes |
| ENSG00000163655 | GMPS | 100 | 88.745 | ENSXETG00000033049 | GMPS | 100 | 88.745 | Xenopus_tropicalis |
| ENSG00000163655 | GMPS | 100 | 85.859 | ENSXMAG00000017018 | gmps | 100 | 85.714 | Xiphophorus_maculatus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0003921 | GMP synthase activity | 21873635. | IBA | Function |
| GO:0003922 | GMP synthase (glutamine-hydrolyzing) activity | 7706277. | EXP | Function |
| GO:0003922 | GMP synthase (glutamine-hydrolyzing) activity | 21873635. | IBA | Function |
| GO:0005524 | ATP binding | - | IEA | Function |
| GO:0005829 | cytosol | 21873635. | IBA | Component |
| GO:0005829 | cytosol | - | IDA | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0006177 | GMP biosynthetic process | 21873635. | IBA | Process |
| GO:0006177 | GMP biosynthetic process | - | IEA | Process |
| GO:0006541 | glutamine metabolic process | - | IEA | Process |
| GO:0009113 | purine nucleobase biosynthetic process | 8089153. | TAS | Process |
| GO:0009168 | purine ribonucleoside monophosphate biosynthetic process | - | TAS | Process |
| GO:0016462 | pyrophosphatase activity | - | IEA | Function |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| ACC | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRP | |||||||
| LAML | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| MESO | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS | |||||||
| UCS |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| KIRC | |||
| KIRP | |||
| PAAD |
| Cancer | P-value | Q-value |
|---|---|---|
| SARC | ||
| MESO | ||
| ACC | ||
| LUSC | ||
| KIRP | ||
| COAD | ||
| PAAD | ||
| PCPG | ||
| CESC | ||
| READ | ||
| KICH | ||
| UCEC | ||
| LIHC | ||
| DLBC | ||
| THCA | ||
| LUAD | ||
| UVM | ||
| OV |