
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000324817 | RAP | PF08373.10 | 1.1e-06 | 1 | 1 |
| ENSP00000418516 | RAP | PF08373.10 | 1.5e-06 | 1 | 1 |
| ENSP00000297532 | RAP | PF08373.10 | 1.6e-06 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000460980 | FASTK-204 | 1521 | - | - | - (aa) | - | - |
| ENST00000478477 | FASTK-210 | 826 | - | - | - (aa) | - | - |
| ENST00000483953 | FASTK-214 | 1178 | - | - | - (aa) | - | - |
| ENST00000467237 | FASTK-208 | 3011 | - | - | - (aa) | - | - |
| ENST00000482571 | FASTK-212 | 1764 | XM_017011704 | ENSP00000418516 | 522 (aa) | XP_016867193 | Q14296 |
| ENST00000465272 | FASTK-206 | 457 | - | - | - (aa) | - | - |
| ENST00000540185 | FASTK-217 | 1609 | XM_011515761 | ENSP00000444498 | 465 (aa) | XP_011514063 | G3V1R6 |
| ENST00000482806 | FASTK-213 | 1826 | - | - | - (aa) | - | - |
| ENST00000489884 | FASTK-215 | 1751 | - | - | - (aa) | - | - |
| ENST00000466855 | FASTK-207 | 1894 | - | - | - (aa) | - | - |
| ENST00000461979 | FASTK-205 | 720 | - | - | - (aa) | - | - |
| ENST00000459800 | FASTK-203 | 449 | - | - | - (aa) | - | - |
| ENST00000469237 | FASTK-209 | 1904 | - | - | - (aa) | - | - |
| ENST00000496663 | FASTK-216 | 818 | - | - | - (aa) | - | - |
| ENST00000353841 | FASTK-202 | 1402 | XM_011515763 | ENSP00000324817 | 408 (aa) | XP_011514065 | Q14296 |
| ENST00000478883 | FASTK-211 | 706 | - | - | - (aa) | - | - |
| ENST00000297532 | FASTK-201 | 1827 | XM_005249932 | ENSP00000297532 | 549 (aa) | XP_005249989 | Q14296 |
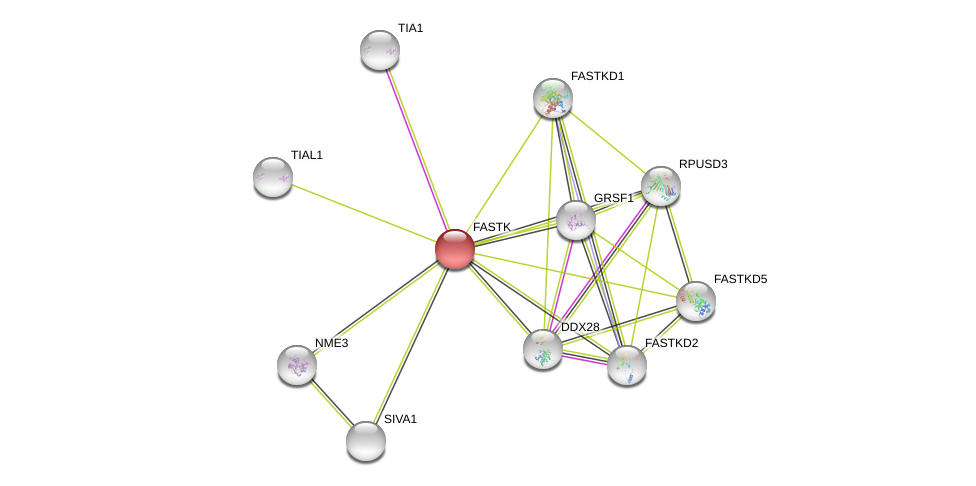
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000164896 | FASTK | 100 | 95.263 | ENSAMEG00000013695 | FASTK | 100 | 93.989 | Ailuropoda_melanoleuca |
| ENSG00000164896 | FASTK | 86 | 44.565 | ENSACIG00000003041 | fastk | 93 | 44.565 | Amphilophus_citrinellus |
| ENSG00000164896 | FASTK | 65 | 47.212 | ENSACAG00000005313 | - | 99 | 47.212 | Anolis_carolinensis |
| ENSG00000164896 | FASTK | 100 | 95.833 | ENSANAG00000028784 | FASTK | 100 | 95.833 | Aotus_nancymaae |
| ENSG00000164896 | FASTK | 82 | 47.619 | ENSACLG00000016505 | fastk | 58 | 47.619 | Astatotilapia_calliptera |
| ENSG00000164896 | FASTK | 100 | 91.053 | ENSBTAG00000011228 | FASTK | 100 | 90.727 | Bos_taurus |
| ENSG00000164896 | FASTK | 100 | 97.059 | ENSCJAG00000020307 | FASTK | 100 | 97.059 | Callithrix_jacchus |
| ENSG00000164896 | FASTK | 100 | 95.263 | ENSCAFG00000004739 | FASTK | 100 | 93.989 | Canis_familiaris |
| ENSG00000164896 | FASTK | 100 | 95.263 | ENSCAFG00020006312 | FASTK | 100 | 93.989 | Canis_lupus_dingo |
| ENSG00000164896 | FASTK | 99 | 94.280 | ENSTSYG00000007984 | FASTK | 99 | 94.280 | Carlito_syrichta |
| ENSG00000164896 | FASTK | 100 | 96.814 | ENSCCAG00000013285 | FASTK | 100 | 96.814 | Cebus_capucinus |
| ENSG00000164896 | FASTK | 100 | 98.947 | ENSCATG00000042475 | FASTK | 100 | 98.947 | Cercocebus_atys |
| ENSG00000164896 | FASTK | 95 | 90.403 | ENSCLAG00000004011 | FASTK | 100 | 90.403 | Chinchilla_lanigera |
| ENSG00000164896 | FASTK | 100 | 98.947 | ENSCSAG00000006690 | FASTK | 100 | 98.179 | Chlorocebus_sabaeus |
| ENSG00000164896 | FASTK | 82 | 56.481 | ENSCPBG00000012219 | FASTK | 51 | 53.226 | Chrysemys_picta_bellii |
| ENSG00000164896 | FASTK | 100 | 98.775 | ENSCANG00000040479 | FASTK | 100 | 98.775 | Colobus_angolensis_palliatus |
| ENSG00000164896 | FASTK | 100 | 89.474 | ENSCGRG00001002508 | Fastk | 100 | 89.474 | Cricetulus_griseus_chok1gshd |
| ENSG00000164896 | FASTK | 95 | 88.676 | ENSCGRG00000012561 | Fastk | 100 | 88.676 | Cricetulus_griseus_crigri |
| ENSG00000164896 | FASTK | 74 | 51.152 | ENSCSEG00000014579 | fastk | 61 | 46.269 | Cynoglossus_semilaevis |
| ENSG00000164896 | FASTK | 82 | 50.190 | ENSDARG00000077075 | fastk | 51 | 50.382 | Danio_rerio |
| ENSG00000164896 | FASTK | 100 | 92.714 | ENSDNOG00000032359 | FASTK | 100 | 92.714 | Dasypus_novemcinctus |
| ENSG00000164896 | FASTK | 100 | 89.071 | ENSDORG00000006552 | Fastk | 100 | 89.071 | Dipodomys_ordii |
| ENSG00000164896 | FASTK | 78 | 49.275 | ENSEBUG00000004770 | fastk | 50 | 49.275 | Eptatretus_burgeri |
| ENSG00000164896 | FASTK | 100 | 96.316 | ENSEASG00005002707 | FASTK | 100 | 95.082 | Equus_asinus_asinus |
| ENSG00000164896 | FASTK | 100 | 96.316 | ENSECAG00000014380 | FASTK | 100 | 95.082 | Equus_caballus |
| ENSG00000164896 | FASTK | 82 | 50.000 | ENSELUG00000017385 | fastk | 90 | 50.000 | Esox_lucius |
| ENSG00000164896 | FASTK | 100 | 95.789 | ENSFCAG00000013198 | FASTK | 100 | 95.789 | Felis_catus |
| ENSG00000164896 | FASTK | 91 | 49.202 | ENSFALG00000013118 | FASTK | 100 | 47.934 | Ficedula_albicollis |
| ENSG00000164896 | FASTK | 82 | 50.602 | ENSFHEG00000021968 | fastk | 79 | 43.908 | Fundulus_heteroclitus |
| ENSG00000164896 | FASTK | 90 | 45.889 | ENSGMOG00000009461 | fastk | 95 | 44.928 | Gadus_morhua |
| ENSG00000164896 | FASTK | 74 | 58.333 | ENSGALG00000033058 | FASTK | 52 | 54.839 | Gallus_gallus |
| ENSG00000164896 | FASTK | 90 | 46.400 | ENSGACG00000001499 | fastk | 99 | 43.789 | Gasterosteus_aculeatus |
| ENSG00000164896 | FASTK | 74 | 57.870 | ENSGAGG00000016922 | FASTK | 52 | 54.435 | Gopherus_agassizii |
| ENSG00000164896 | FASTK | 100 | 99.818 | ENSGGOG00000016906 | FASTK | 100 | 99.818 | Gorilla_gorilla |
| ENSG00000164896 | FASTK | 82 | 47.619 | ENSHBUG00000009697 | fastk | 54 | 47.619 | Haplochromis_burtoni |
| ENSG00000164896 | FASTK | 97 | 64.750 | ENSHGLG00000003297 | FASTK | 100 | 64.750 | Heterocephalus_glaber_female |
| ENSG00000164896 | FASTK | 100 | 64.144 | ENSHGLG00100015402 | FASTK | 100 | 64.144 | Heterocephalus_glaber_male |
| ENSG00000164896 | FASTK | 78 | 53.125 | ENSHCOG00000013053 | fastk | 51 | 50.000 | Hippocampus_comes |
| ENSG00000164896 | FASTK | 75 | 54.839 | ENSIPUG00000002527 | FASTK | 50 | 51.406 | Ictalurus_punctatus |
| ENSG00000164896 | FASTK | 100 | 97.105 | ENSSTOG00000016075 | FASTK | 100 | 97.105 | Ictidomys_tridecemlineatus |
| ENSG00000164896 | FASTK | 100 | 88.537 | ENSJJAG00000016554 | Fastk | 100 | 88.537 | Jaculus_jaculus |
| ENSG00000164896 | FASTK | 78 | 49.221 | ENSLACG00000010865 | FASTK | 100 | 49.216 | Latimeria_chalumnae |
| ENSG00000164896 | FASTK | 82 | 47.619 | ENSLOCG00000010214 | fastk | 51 | 47.619 | Lepisosteus_oculatus |
| ENSG00000164896 | FASTK | 100 | 91.621 | ENSLAFG00000008796 | FASTK | 100 | 91.621 | Loxodonta_africana |
| ENSG00000164896 | FASTK | 100 | 99.020 | ENSMFAG00000041547 | FASTK | 100 | 99.020 | Macaca_fascicularis |
| ENSG00000164896 | FASTK | 100 | 98.947 | ENSMMUG00000020459 | FASTK | 100 | 98.947 | Macaca_mulatta |
| ENSG00000164896 | FASTK | 100 | 99.020 | ENSMNEG00000008431 | FASTK | 100 | 99.020 | Macaca_nemestrina |
| ENSG00000164896 | FASTK | 95 | 98.273 | ENSMLEG00000029888 | FASTK | 100 | 98.273 | Mandrillus_leucophaeus |
| ENSG00000164896 | FASTK | 82 | 47.619 | ENSMZEG00005019400 | fastk | 58 | 47.619 | Maylandia_zebra |
| ENSG00000164896 | FASTK | 100 | 88.684 | ENSMAUG00000015620 | Fastk | 100 | 88.684 | Mesocricetus_auratus |
| ENSG00000164896 | FASTK | 100 | 95.833 | ENSMICG00000004757 | FASTK | 100 | 95.833 | Microcebus_murinus |
| ENSG00000164896 | FASTK | 94 | 88.544 | ENSMOCG00000020800 | Fastk | 100 | 88.544 | Microtus_ochrogaster |
| ENSG00000164896 | FASTK | 74 | 46.774 | ENSMMOG00000018047 | fastk | 56 | 46.774 | Mola_mola |
| ENSG00000164896 | FASTK | 83 | 45.022 | ENSMALG00000000220 | fastk | 86 | 42.740 | Monopterus_albus |
| ENSG00000164896 | FASTK | 100 | 89.461 | MGP_CAROLIEiJ_G0027012 | Fastk | 100 | 90.000 | Mus_caroli |
| ENSG00000164896 | FASTK | 100 | 89.461 | ENSMUSG00000028959 | Fastk | 100 | 90.000 | Mus_musculus |
| ENSG00000164896 | FASTK | 100 | 90.000 | MGP_PahariEiJ_G0021785 | Fastk | 100 | 90.000 | Mus_pahari |
| ENSG00000164896 | FASTK | 100 | 89.737 | MGP_SPRETEiJ_G0028008 | Fastk | 100 | 89.737 | Mus_spretus |
| ENSG00000164896 | FASTK | 100 | 93.684 | ENSMPUG00000008727 | FASTK | 100 | 92.714 | Mustela_putorius_furo |
| ENSG00000164896 | FASTK | 100 | 91.842 | ENSMLUG00000014730 | FASTK | 100 | 90.710 | Myotis_lucifugus |
| ENSG00000164896 | FASTK | 100 | 92.157 | ENSNGAG00000010690 | Fastk | 100 | 92.157 | Nannospalax_galili |
| ENSG00000164896 | FASTK | 82 | 47.619 | ENSNBRG00000017186 | fastk | 54 | 47.619 | Neolamprologus_brichardi |
| ENSG00000164896 | FASTK | 100 | 99.020 | ENSNLEG00000004192 | FASTK | 100 | 99.020 | Nomascus_leucogenys |
| ENSG00000164896 | FASTK | 77 | 94.161 | ENSOPRG00000003885 | FASTK | 68 | 94.161 | Ochotona_princeps |
| ENSG00000164896 | FASTK | 100 | 88.725 | ENSODEG00000015168 | FASTK | 100 | 88.725 | Octodon_degus |
| ENSG00000164896 | FASTK | 90 | 46.702 | ENSONIG00000014035 | fastk | 100 | 43.066 | Oreochromis_niloticus |
| ENSG00000164896 | FASTK | 100 | 95.789 | ENSOCUG00000023964 | FASTK | 100 | 94.353 | Oryctolagus_cuniculus |
| ENSG00000164896 | FASTK | 75 | 55.046 | ENSORLG00000023667 | fastk | 51 | 48.299 | Oryzias_latipes |
| ENSG00000164896 | FASTK | 84 | 47.782 | ENSORLG00020018249 | fastk | 50 | 47.945 | Oryzias_latipes_hni |
| ENSG00000164896 | FASTK | 77 | 53.879 | ENSORLG00015016956 | fastk | 50 | 50.951 | Oryzias_latipes_hsok |
| ENSG00000164896 | FASTK | 82 | 48.913 | ENSOMEG00000022058 | fastk | 51 | 48.913 | Oryzias_melastigma |
| ENSG00000164896 | FASTK | 100 | 96.316 | ENSOGAG00000001782 | FASTK | 100 | 94.536 | Otolemur_garnettii |
| ENSG00000164896 | FASTK | 100 | 99.636 | ENSPPAG00000042376 | FASTK | 100 | 99.636 | Pan_paniscus |
| ENSG00000164896 | FASTK | 100 | 96.053 | ENSPPRG00000003158 | FASTK | 100 | 96.053 | Panthera_pardus |
| ENSG00000164896 | FASTK | 93 | 96.053 | ENSPTIG00000012926 | FASTK | 100 | 94.175 | Panthera_tigris_altaica |
| ENSG00000164896 | FASTK | 100 | 99.818 | ENSPTRG00000019874 | FASTK | 100 | 99.818 | Pan_troglodytes |
| ENSG00000164896 | FASTK | 100 | 99.020 | ENSPANG00000015942 | FASTK | 100 | 99.020 | Papio_anubis |
| ENSG00000164896 | FASTK | 82 | 47.703 | ENSPKIG00000008408 | fastk | 52 | 47.872 | Paramormyrops_kingsleyae |
| ENSG00000164896 | FASTK | 81 | 40.889 | ENSPMGG00000003684 | fastk | 84 | 41.204 | Periophthalmus_magnuspinnatus |
| ENSG00000164896 | FASTK | 100 | 89.211 | ENSPEMG00000012621 | Fastk | 100 | 89.211 | Peromyscus_maniculatus_bairdii |
| ENSG00000164896 | FASTK | 90 | 42.359 | ENSPMAG00000009209 | fastk | 100 | 41.087 | Petromyzon_marinus |
| ENSG00000164896 | FASTK | 78 | 50.800 | ENSPFOG00000010879 | fastk | 52 | 51.004 | Poecilia_formosa |
| ENSG00000164896 | FASTK | 81 | 50.382 | ENSPLAG00000016993 | fastk | 53 | 48.421 | Poecilia_latipinna |
| ENSG00000164896 | FASTK | 79 | 50.800 | ENSPMEG00000009803 | fastk | 51 | 51.004 | Poecilia_mexicana |
| ENSG00000164896 | FASTK | 79 | 50.800 | ENSPREG00000001443 | fastk | 51 | 51.004 | Poecilia_reticulata |
| ENSG00000164896 | FASTK | 100 | 99.271 | ENSPPYG00000018185 | FASTK | 100 | 99.271 | Pongo_abelii |
| ENSG00000164896 | FASTK | 100 | 93.684 | ENSPCAG00000007566 | FASTK | 100 | 92.350 | Procavia_capensis |
| ENSG00000164896 | FASTK | 100 | 96.053 | ENSPCOG00000028719 | FASTK | 100 | 96.053 | Propithecus_coquereli |
| ENSG00000164896 | FASTK | 91 | 94.921 | ENSPVAG00000007095 | FASTK | 88 | 95.238 | Pteropus_vampyrus |
| ENSG00000164896 | FASTK | 82 | 47.619 | ENSPNYG00000004170 | fastk | 58 | 47.619 | Pundamilia_nyererei |
| ENSG00000164896 | FASTK | 100 | 98.421 | ENSRBIG00000030083 | FASTK | 100 | 98.421 | Rhinopithecus_bieti |
| ENSG00000164896 | FASTK | 100 | 98.421 | ENSRROG00000042586 | FASTK | 100 | 98.421 | Rhinopithecus_roxellana |
| ENSG00000164896 | FASTK | 93 | 96.579 | ENSSBOG00000033161 | FASTK | 100 | 95.146 | Saimiri_boliviensis_boliviensis |
| ENSG00000164896 | FASTK | 82 | 45.397 | ENSSFOG00015019472 | fastk | 59 | 45.397 | Scleropages_formosus |
| ENSG00000164896 | FASTK | 76 | 53.670 | ENSSDUG00000006952 | fastk | 52 | 51.406 | Seriola_dumerili |
| ENSG00000164896 | FASTK | 92 | 41.505 | ENSSPUG00000006560 | FASTK | 67 | 56.522 | Sphenodon_punctatus |
| ENSG00000164896 | FASTK | 82 | 48.387 | ENSSPAG00000020341 | fastk | 85 | 44.098 | Stegastes_partitus |
| ENSG00000164896 | FASTK | 100 | 91.842 | ENSSSCG00000024184 | FASTK | 100 | 91.439 | Sus_scrofa |
| ENSG00000164896 | FASTK | 82 | 47.260 | ENSTRUG00000023502 | fastk | 52 | 46.552 | Takifugu_rubripes |
| ENSG00000164896 | FASTK | 93 | 44.560 | ENSTNIG00000003067 | fastk | 99 | 49.383 | Tetraodon_nigroviridis |
| ENSG00000164896 | FASTK | 100 | 91.579 | ENSTTRG00000000853 | FASTK | 100 | 91.075 | Tursiops_truncatus |
| ENSG00000164896 | FASTK | 100 | 95.789 | ENSUAMG00000022753 | FASTK | 100 | 95.789 | Ursus_americanus |
| ENSG00000164896 | FASTK | 95 | 94.818 | ENSUMAG00000016282 | FASTK | 97 | 94.818 | Ursus_maritimus |
| ENSG00000164896 | FASTK | 71 | 92.617 | ENSVPAG00000006535 | FASTK | 75 | 92.617 | Vicugna_pacos |
| ENSG00000164896 | FASTK | 100 | 95.343 | ENSVVUG00000008400 | FASTK | 100 | 95.343 | Vulpes_vulpes |
| ENSG00000164896 | FASTK | 75 | 49.518 | ENSXCOG00000000382 | fastk | 78 | 49.838 | Xiphophorus_couchianus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0003723 | RNA binding | - | IEA | Function |
| GO:0004674 | protein serine/threonine kinase activity | 7544399. | IDA | Function |
| GO:0005515 | protein binding | 7544399.15110758.25416956.25910212. | IPI | Function |
| GO:0005524 | ATP binding | - | IEA | Function |
| GO:0005759 | mitochondrial matrix | - | IEA | Component |
| GO:0006468 | protein phosphorylation | 7544399. | IDA | Process |
| GO:0033867 | Fas-activated serine/threonine kinase activity | - | IEA | Function |
| GO:0043484 | regulation of RNA splicing | 17135269. | IDA | Process |
| GO:0043484 | regulation of RNA splicing | 17592127. | IMP | Process |
| GO:0097190 | apoptotic signaling pathway | 7544399. | TAS | Process |