
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000440054 | zf-C2H2 | PF00096.26 | 1.1e-17 | 1 | 4 |
| ENSP00000440054 | zf-C2H2 | PF00096.26 | 1.1e-17 | 2 | 4 |
| ENSP00000440054 | zf-C2H2 | PF00096.26 | 1.1e-17 | 3 | 4 |
| ENSP00000440054 | zf-C2H2 | PF00096.26 | 1.1e-17 | 4 | 4 |
| ENSP00000396773 | zf-C2H2 | PF00096.26 | 1.2e-17 | 1 | 4 |
| ENSP00000396773 | zf-C2H2 | PF00096.26 | 1.2e-17 | 2 | 4 |
| ENSP00000396773 | zf-C2H2 | PF00096.26 | 1.2e-17 | 3 | 4 |
| ENSP00000396773 | zf-C2H2 | PF00096.26 | 1.2e-17 | 4 | 4 |
| ENSP00000483562 | zf-C2H2 | PF00096.26 | 2.9e-17 | 1 | 4 |
| ENSP00000483562 | zf-C2H2 | PF00096.26 | 2.9e-17 | 2 | 4 |
| ENSP00000483562 | zf-C2H2 | PF00096.26 | 2.9e-17 | 3 | 4 |
| ENSP00000483562 | zf-C2H2 | PF00096.26 | 2.9e-17 | 4 | 4 |
| ENSP00000440054 | zf-met | PF12874.7 | 1.9e-11 | 1 | 2 |
| ENSP00000440054 | zf-met | PF12874.7 | 1.9e-11 | 2 | 2 |
| ENSP00000396773 | zf-met | PF12874.7 | 1.9e-11 | 1 | 2 |
| ENSP00000396773 | zf-met | PF12874.7 | 1.9e-11 | 2 | 2 |
| ENSP00000483562 | zf-met | PF12874.7 | 4.6e-11 | 1 | 2 |
| ENSP00000483562 | zf-met | PF12874.7 | 4.6e-11 | 2 | 2 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000614342 | SALL2-207 | 4942 | - | ENSP00000483562 | 1007 (aa) | - | Q9Y467 |
| ENST00000613414 | SALL2-206 | 1566 | - | ENSP00000483202 | 200 (aa) | - | A0A0B4J2F7 |
| ENST00000546363 | SALL2-204 | 3094 | XM_011537064 | ENSP00000440054 | 902 (aa) | XP_011535366 | H0YFS7 |
| ENST00000537235 | SALL2-202 | 659 | - | ENSP00000438493 | 191 (aa) | - | F5H433 |
| ENST00000450879 | SALL2-201 | 3178 | XM_011537065 | ENSP00000396773 | 906 (aa) | XP_011535367 | E7EW59 |
| ENST00000541965 | SALL2-203 | 553 | - | ENSP00000439654 | 99 (aa) | - | F5H1G6 |
| ENST00000611430 | SALL2-205 | 1466 | - | ENSP00000484460 | 198 (aa) | - | A0A087X1T9 |
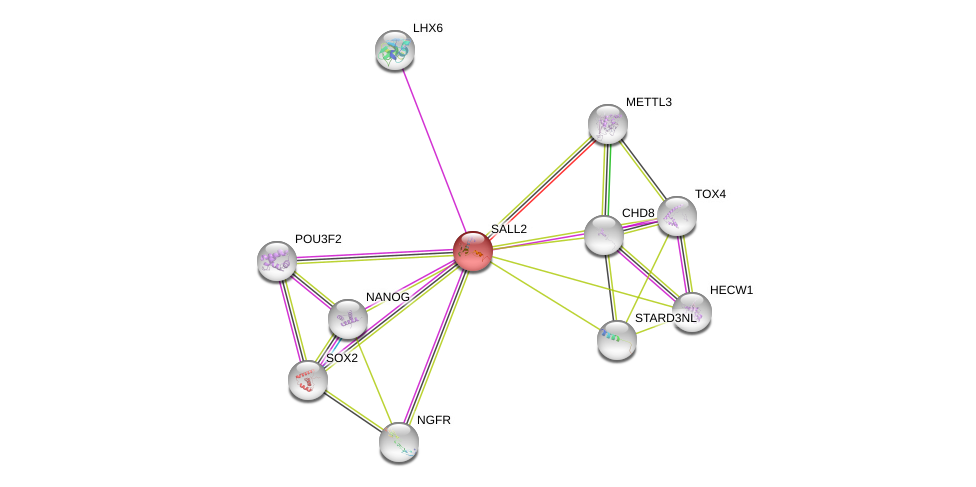
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000165821 | SALL2 | 100 | 90.909 | ENSAMEG00000006226 | SALL2 | 96 | 90.415 | Ailuropoda_melanoleuca |
| ENSG00000165821 | SALL2 | 63 | 60.202 | ENSACAG00000000030 | SALL2 | 76 | 60.371 | Anolis_carolinensis |
| ENSG00000165821 | SALL2 | 100 | 97.382 | ENSANAG00000031405 | SALL2 | 90 | 96.000 | Aotus_nancymaae |
| ENSG00000165821 | SALL2 | 100 | 91.099 | ENSBTAG00000013249 | SALL2 | 99 | 90.707 | Bos_taurus |
| ENSG00000165821 | SALL2 | 100 | 98.990 | ENSCJAG00000008855 | SALL2 | 100 | 97.621 | Callithrix_jacchus |
| ENSG00000165821 | SALL2 | 89 | 73.864 | ENSCAFG00000005676 | SALL2 | 95 | 73.913 | Canis_familiaris |
| ENSG00000165821 | SALL2 | 88 | 83.529 | ENSCAFG00020015273 | SALL2 | 100 | 77.044 | Canis_lupus_dingo |
| ENSG00000165821 | SALL2 | 100 | 90.864 | ENSCHIG00000025803 | SALL2 | 100 | 90.765 | Capra_hircus |
| ENSG00000165821 | SALL2 | 98 | 95.467 | ENSTSYG00000030220 | SALL2 | 99 | 92.746 | Carlito_syrichta |
| ENSG00000165821 | SALL2 | 100 | 90.217 | ENSCPOG00000040220 | SALL2 | 100 | 88.142 | Cavia_porcellus |
| ENSG00000165821 | SALL2 | 100 | 97.324 | ENSCCAG00000033278 | SALL2 | 100 | 97.324 | Cebus_capucinus |
| ENSG00000165821 | SALL2 | 100 | 100.000 | ENSCATG00000045263 | SALL2 | 100 | 98.811 | Cercocebus_atys |
| ENSG00000165821 | SALL2 | 100 | 90.099 | ENSCLAG00000001400 | SALL2 | 100 | 88.515 | Chinchilla_lanigera |
| ENSG00000165821 | SALL2 | 100 | 93.936 | ENSCSAG00000016461 | SALL2 | 100 | 93.936 | Chlorocebus_sabaeus |
| ENSG00000165821 | SALL2 | 85 | 52.750 | ENSCPBG00000012695 | SALL2 | 61 | 68.007 | Chrysemys_picta_bellii |
| ENSG00000165821 | SALL2 | 100 | 98.990 | ENSCANG00000036872 | SALL2 | 100 | 98.513 | Colobus_angolensis_palliatus |
| ENSG00000165821 | SALL2 | 100 | 88.194 | ENSCGRG00001000154 | Sall2 | 100 | 87.698 | Cricetulus_griseus_chok1gshd |
| ENSG00000165821 | SALL2 | 100 | 88.095 | ENSCGRG00000008130 | Sall2 | 100 | 87.599 | Cricetulus_griseus_crigri |
| ENSG00000165821 | SALL2 | 100 | 89.529 | ENSDNOG00000032317 | SALL2 | 99 | 90.515 | Dasypus_novemcinctus |
| ENSG00000165821 | SALL2 | 100 | 88.987 | ENSDORG00000030154 | Sall2 | 100 | 88.594 | Dipodomys_ordii |
| ENSG00000165821 | SALL2 | 100 | 95.288 | ENSEASG00005011556 | SALL2 | 100 | 93.168 | Equus_asinus_asinus |
| ENSG00000165821 | SALL2 | 94 | 93.565 | ENSECAG00000024449 | SALL2 | 100 | 93.565 | Equus_caballus |
| ENSG00000165821 | SALL2 | 100 | 91.601 | ENSFCAG00000044786 | SALL2 | 100 | 91.502 | Felis_catus |
| ENSG00000165821 | SALL2 | 100 | 91.790 | ENSFDAG00000014250 | SALL2 | 100 | 89.812 | Fukomys_damarensis |
| ENSG00000165821 | SALL2 | 100 | 100.000 | ENSGGOG00000009133 | SALL2 | 100 | 99.206 | Gorilla_gorilla |
| ENSG00000165821 | SALL2 | 100 | 92.857 | ENSHGLG00000006009 | SALL2 | 100 | 91.071 | Heterocephalus_glaber_female |
| ENSG00000165821 | SALL2 | 98 | 92.417 | ENSHGLG00100000859 | SALL2 | 99 | 90.719 | Heterocephalus_glaber_male |
| ENSG00000165821 | SALL2 | 100 | 93.452 | ENSSTOG00000023637 | SALL2 | 100 | 92.460 | Ictidomys_tridecemlineatus |
| ENSG00000165821 | SALL2 | 100 | 88.702 | ENSJJAG00000000077 | Sall2 | 100 | 87.711 | Jaculus_jaculus |
| ENSG00000165821 | SALL2 | 100 | 92.864 | ENSLAFG00000032150 | SALL2 | 100 | 91.774 | Loxodonta_africana |
| ENSG00000165821 | SALL2 | 100 | 100.000 | ENSMFAG00000002687 | SALL2 | 100 | 98.811 | Macaca_fascicularis |
| ENSG00000165821 | SALL2 | 100 | 98.236 | ENSMMUG00000014997 | SALL2 | 100 | 98.236 | Macaca_mulatta |
| ENSG00000165821 | SALL2 | 100 | 100.000 | ENSMNEG00000030415 | SALL2 | 100 | 98.346 | Macaca_nemestrina |
| ENSG00000165821 | SALL2 | 100 | 100.000 | ENSMLEG00000036462 | SALL2 | 100 | 98.910 | Mandrillus_leucophaeus |
| ENSG00000165821 | SALL2 | 100 | 88.142 | ENSMAUG00000013964 | Sall2 | 100 | 86.957 | Mesocricetus_auratus |
| ENSG00000165821 | SALL2 | 100 | 97.980 | ENSMICG00000046438 | SALL2 | 100 | 93.247 | Microcebus_murinus |
| ENSG00000165821 | SALL2 | 100 | 86.905 | ENSMOCG00000003410 | Sall2 | 100 | 86.111 | Microtus_ochrogaster |
| ENSG00000165821 | SALL2 | 99 | 79.277 | ENSMODG00000006068 | SALL2 | 99 | 78.495 | Monodelphis_domestica |
| ENSG00000165821 | SALL2 | 100 | 88.381 | MGP_CAROLIEiJ_G0019241 | Sall2 | 100 | 88.083 | Mus_caroli |
| ENSG00000165821 | SALL2 | 100 | 90.052 | ENSMUSG00000049532 | Sall2 | 100 | 88.282 | Mus_musculus |
| ENSG00000165821 | SALL2 | 100 | 88.679 | MGP_PahariEiJ_G0030262 | Sall2 | 100 | 87.885 | Mus_pahari |
| ENSG00000165821 | SALL2 | 100 | 88.580 | MGP_SPRETEiJ_G0020133 | Sall2 | 100 | 88.282 | Mus_spretus |
| ENSG00000165821 | SALL2 | 100 | 93.194 | ENSMPUG00000000199 | SALL2 | 88 | 89.922 | Mustela_putorius_furo |
| ENSG00000165821 | SALL2 | 100 | 91.179 | ENSMLUG00000009185 | SALL2 | 99 | 91.179 | Myotis_lucifugus |
| ENSG00000165821 | SALL2 | 100 | 90.278 | ENSNGAG00000014258 | Sall2 | 100 | 89.385 | Nannospalax_galili |
| ENSG00000165821 | SALL2 | 100 | 100.000 | ENSNLEG00000014602 | SALL2 | 100 | 98.611 | Nomascus_leucogenys |
| ENSG00000165821 | SALL2 | 91 | 79.871 | ENSMEUG00000000526 | SALL2 | 100 | 78.902 | Notamacropus_eugenii |
| ENSG00000165821 | SALL2 | 100 | 90.387 | ENSODEG00000001454 | SALL2 | 100 | 89.197 | Octodon_degus |
| ENSG00000165821 | SALL2 | 100 | 88.137 | ENSOCUG00000027464 | SALL2 | 100 | 89.258 | Oryctolagus_cuniculus |
| ENSG00000165821 | SALL2 | 100 | 97.980 | ENSOGAG00000031464 | SALL2 | 100 | 93.162 | Otolemur_garnettii |
| ENSG00000165821 | SALL2 | 100 | 90.070 | ENSOARG00000019612 | SALL2 | 100 | 89.970 | Ovis_aries |
| ENSG00000165821 | SALL2 | 100 | 100.000 | ENSPPAG00000038242 | SALL2 | 100 | 99.503 | Pan_paniscus |
| ENSG00000165821 | SALL2 | 100 | 92.079 | ENSPPRG00000005572 | SALL2 | 100 | 91.980 | Panthera_pardus |
| ENSG00000165821 | SALL2 | 100 | 89.296 | ENSPTIG00000018399 | SALL2 | 100 | 89.197 | Panthera_tigris_altaica |
| ENSG00000165821 | SALL2 | 100 | 100.000 | ENSPTRG00000006128 | SALL2 | 100 | 99.305 | Pan_troglodytes |
| ENSG00000165821 | SALL2 | 100 | 98.990 | ENSPANG00000020006 | SALL2 | 100 | 98.414 | Papio_anubis |
| ENSG00000165821 | SALL2 | 100 | 89.911 | ENSPEMG00000022293 | Sall2 | 100 | 88.922 | Peromyscus_maniculatus_bairdii |
| ENSG00000165821 | SALL2 | 100 | 99.200 | ENSPPYG00000005600 | SALL2 | 92 | 97.799 | Pongo_abelii |
| ENSG00000165821 | SALL2 | 91 | 91.667 | ENSPCAG00000006689 | SALL2 | 89 | 89.444 | Procavia_capensis |
| ENSG00000165821 | SALL2 | 100 | 93.843 | ENSPCOG00000017266 | SALL2 | 100 | 93.049 | Propithecus_coquereli |
| ENSG00000165821 | SALL2 | 100 | 88.107 | ENSPVAG00000000277 | SALL2 | 100 | 87.909 | Pteropus_vampyrus |
| ENSG00000165821 | SALL2 | 100 | 89.076 | ENSRNOG00000013287 | Sall2 | 100 | 88.282 | Rattus_norvegicus |
| ENSG00000165821 | SALL2 | 87 | 98.082 | ENSRBIG00000009332 | SALL2 | 100 | 100.000 | Rhinopithecus_bieti |
| ENSG00000165821 | SALL2 | 100 | 88.999 | ENSRROG00000035352 | SALL2 | 95 | 95.524 | Rhinopithecus_roxellana |
| ENSG00000165821 | SALL2 | 100 | 97.906 | ENSSBOG00000028539 | SALL2 | 100 | 97.423 | Saimiri_boliviensis_boliviensis |
| ENSG00000165821 | SALL2 | 100 | 74.872 | ENSSHAG00000009219 | SALL2 | 98 | 76.547 | Sarcophilus_harrisii |
| ENSG00000165821 | SALL2 | 59 | 68.223 | ENSSPUG00000010045 | SALL2 | 58 | 68.571 | Sphenodon_punctatus |
| ENSG00000165821 | SALL2 | 99 | 92.000 | ENSSSCG00000022705 | SALL2 | 91 | 92.000 | Sus_scrofa |
| ENSG00000165821 | SALL2 | 100 | 90.367 | ENSTTRG00000012842 | SALL2 | 100 | 90.169 | Tursiops_truncatus |
| ENSG00000165821 | SALL2 | 91 | 91.111 | ENSUAMG00000028082 | SALL2 | 88 | 88.406 | Ursus_americanus |
| ENSG00000165821 | SALL2 | 94 | 93.388 | ENSUMAG00000004963 | SALL2 | 100 | 86.067 | Ursus_maritimus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000122 | negative regulation of transcription by RNA polymerase II | 21873635. | IBA | Process |
| GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding | 21362508. | IDA | Function |
| GO:0000981 | DNA-binding transcription factor activity, RNA polymerase II-specific | - | ISA | Function |
| GO:0000981 | DNA-binding transcription factor activity, RNA polymerase II-specific | 19274049. | NAS | Function |
| GO:0001228 | DNA-binding transcription activator activity, RNA polymerase II-specific | 21362508. | IDA | Function |
| GO:0001228 | DNA-binding transcription activator activity, RNA polymerase II-specific | 21362508. | IMP | Function |
| GO:0001654 | eye development | 24412933. | IDA | Process |
| GO:0005515 | protein binding | 16189514.19131967.25416956. | IPI | Function |
| GO:0005634 | nucleus | 21873635. | IBA | Component |
| GO:0006355 | regulation of transcription, DNA-templated | 21873635. | IBA | Process |
| GO:0021915 | neural tube development | - | IEA | Process |
| GO:0044877 | protein-containing complex binding | - | IEA | Function |
| GO:0045944 | positive regulation of transcription by RNA polymerase II | 21362508. | IMP | Process |
| GO:0046872 | metal ion binding | - | IEA | Function |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| ACC | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KICH | |||||||
| KIRP | |||||||
| LAML | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PCPG | |||||||
| PRAD | |||||||
| PRAD | |||||||
| READ | |||||||
| SARC | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| THCA | |||||||
| THCA | |||||||
| THCA | |||||||
| THCA | |||||||
| THYM | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| ACC | |||
| ACC | |||
| CHOL | |||
| HNSC | |||
| KIRP | |||
| LUSC | |||
| PAAD |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| STAD | ||
| ACC | ||
| PAAD | ||
| UCEC | ||
| LIHC | ||
| DLBC | ||
| LGG | ||
| LUAD |