
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000558492 | PPIB-202 | 381 | - | - | - (aa) | - | - |
| ENST00000561048 | PPIB-203 | 1208 | - | - | - (aa) | - | - |
| ENST00000300026 | PPIB-201 | 1081 | - | ENSP00000300026 | 216 (aa) | - | P23284 |
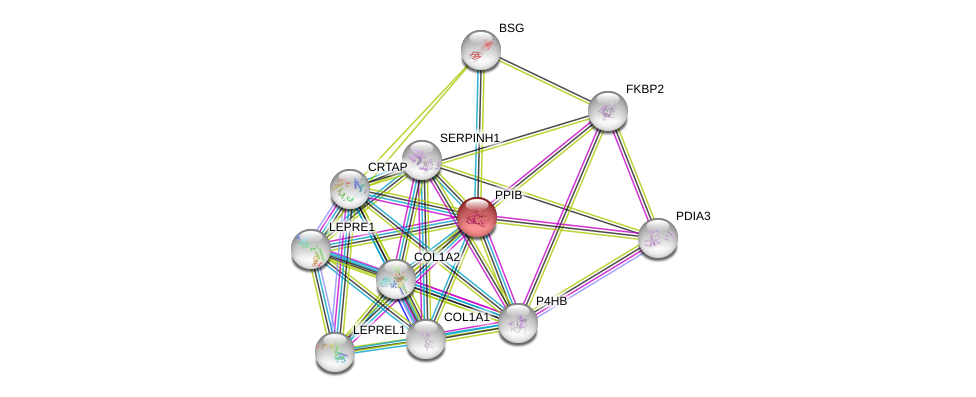
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000166794 | PPIB | 100 | 93.981 | ENSMUSG00000032383 | Ppib | 100 | 93.981 | Mus_musculus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000413 | protein peptidyl-prolyl isomerization | 20676357. | IDA | Process |
| GO:0000413 | protein peptidyl-prolyl isomerization | - | ISS | Process |
| GO:0003723 | RNA binding | 22658674.22681889. | HDA | Function |
| GO:0003755 | peptidyl-prolyl cis-trans isomerase activity | 21873635. | IBA | Function |
| GO:0003755 | peptidyl-prolyl cis-trans isomerase activity | 20676357. | IDA | Function |
| GO:0003755 | peptidyl-prolyl cis-trans isomerase activity | - | ISS | Function |
| GO:0003755 | peptidyl-prolyl cis-trans isomerase activity | 2000394. | NAS | Function |
| GO:0005515 | protein binding | 15095401.15989969.21280149.21516116.22665516.25416956.30021884. | IPI | Function |
| GO:0005515 | protein binding | - | ISS | Function |
| GO:0005518 | collagen binding | 21873635. | IBA | Function |
| GO:0005518 | collagen binding | - | ISS | Function |
| GO:0005634 | nucleus | 21630459. | HDA | Component |
| GO:0005634 | nucleus | - | IDA | Component |
| GO:0005783 | endoplasmic reticulum | 20089953. | IDA | Component |
| GO:0005788 | endoplasmic reticulum lumen | 1530944. | NAS | Component |
| GO:0005788 | endoplasmic reticulum lumen | - | TAS | Component |
| GO:0005790 | smooth endoplasmic reticulum | - | IEA | Component |
| GO:0005925 | focal adhesion | 21423176. | HDA | Component |
| GO:0016018 | cyclosporin A binding | 21873635. | IBA | Function |
| GO:0016018 | cyclosporin A binding | 20676357. | IDA | Function |
| GO:0016020 | membrane | 19946888. | HDA | Component |
| GO:0032991 | protein-containing complex | - | ISS | Component |
| GO:0034663 | endoplasmic reticulum chaperone complex | - | IEA | Component |
| GO:0040018 | positive regulation of multicellular organism growth | 20089953. | IMP | Process |
| GO:0042026 | protein refolding | 21873635. | IBA | Process |
| GO:0042470 | melanosome | - | IEA | Component |
| GO:0044794 | positive regulation by host of viral process | 15989969. | IMP | Process |
| GO:0044829 | positive regulation by host of viral genome replication | 15989969. | IMP | Process |
| GO:0048471 | perinuclear region of cytoplasm | 15989969. | IDA | Component |
| GO:0050821 | protein stabilization | 20089953. | IMP | Process |
| GO:0051082 | unfolded protein binding | 21873635. | IBA | Function |
| GO:0060348 | bone development | 20089953. | IMP | Process |
| GO:0061077 | chaperone-mediated protein folding | 22665516. | IDA | Process |
| GO:0061077 | chaperone-mediated protein folding | 20089953. | IMP | Process |
| GO:0070062 | extracellular exosome | 19056867.19199708.20458337.23533145. | HDA | Component |
| GO:0070063 | RNA polymerase binding | 15989969. | IPI | Function |
| GO:1901873 | regulation of post-translational protein modification | 20089953. | IMP | Process |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| DLBC | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRP | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| THYM | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| KICH | |||
| LGG | |||
| PAAD | |||
| PAAD |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| ACC | ||
| HNSC | ||
| PRAD | ||
| LUSC | ||
| BRCA | ||
| KIRP | ||
| CESC | ||
| KICH | ||
| UCEC | ||
| LGG | ||
| LUAD | ||
| UVM | ||
| OV |