
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000471154 | RIO1 | PF01163.22 | 0.00018 | 1 | 1 |
| ENSP00000472988 | RIO1 | PF01163.22 | 0.00018 | 1 | 1 |
| ENSP00000301264 | RIO1 | PF01163.22 | 0.00067 | 1 | 1 |
| ENSP00000442973 | RIO1 | PF01163.22 | 0.00067 | 1 | 1 |
| PID | Title | Article | Time | Author | Doi |
|---|---|---|---|---|---|
| 29566767 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000601824 | DAPK3-207 | 599 | - | ENSP00000472988 | 158 (aa) | - | M0R0D0 |
| ENST00000594894 | DAPK3-204 | 617 | - | ENSP00000470168 | 92 (aa) | - | M0QYY8 |
| ENST00000595279 | DAPK3-205 | 2058 | - | - | - (aa) | - | - |
| ENST00000593844 | DAPK3-203 | 626 | - | ENSP00000470115 | 141 (aa) | - | M0QYW5 |
| ENST00000301264 | DAPK3-201 | 2105 | - | ENSP00000301264 | 454 (aa) | - | O43293 |
| ENST00000545797 | DAPK3-202 | 2257 | XM_005259508 | ENSP00000442973 | 454 (aa) | XP_005259565 | O43293 |
| ENST00000596311 | DAPK3-206 | 671 | - | ENSP00000471154 | 158 (aa) | - | M0R0D0 |
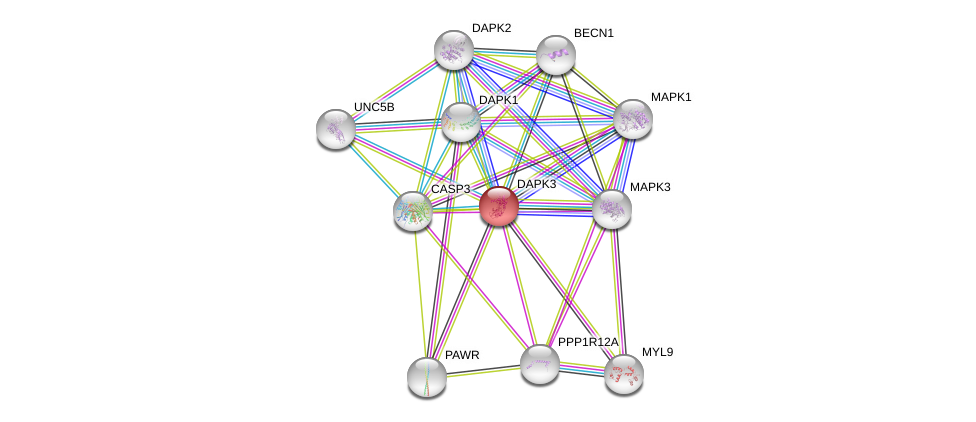
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000167657 | DAPK3 | 100 | 91.139 | ENSAPOG00000005339 | dapk3 | 99 | 80.088 | Acanthochromis_polyacanthus |
| ENSG00000167657 | DAPK3 | 100 | 98.734 | ENSAMEG00000006056 | DAPK3 | 100 | 94.053 | Ailuropoda_melanoleuca |
| ENSG00000167657 | DAPK3 | 100 | 91.139 | ENSACIG00000007416 | dapk3 | 99 | 78.540 | Amphilophus_citrinellus |
| ENSG00000167657 | DAPK3 | 100 | 91.772 | ENSAOCG00000023366 | dapk3 | 99 | 80.752 | Amphiprion_ocellaris |
| ENSG00000167657 | DAPK3 | 100 | 91.772 | ENSAPEG00000021762 | dapk3 | 99 | 80.752 | Amphiprion_percula |
| ENSG00000167657 | DAPK3 | 100 | 99.367 | ENSBTAG00000020417 | DAPK3 | 100 | 94.934 | Bos_taurus |
| ENSG00000167657 | DAPK3 | 100 | 100.000 | ENSCJAG00000015041 | DAPK3 | 100 | 100.000 | Callithrix_jacchus |
| ENSG00000167657 | DAPK3 | 100 | 99.367 | ENSCAFG00000019167 | DAPK3 | 100 | 95.374 | Canis_familiaris |
| ENSG00000167657 | DAPK3 | 100 | 99.367 | ENSCAFG00020018374 | DAPK3 | 100 | 95.374 | Canis_lupus_dingo |
| ENSG00000167657 | DAPK3 | 100 | 99.367 | ENSCHIG00000018026 | DAPK3 | 100 | 94.934 | Capra_hircus |
| ENSG00000167657 | DAPK3 | 100 | 98.101 | ENSTSYG00000035745 | DAPK3 | 100 | 97.857 | Carlito_syrichta |
| ENSG00000167657 | DAPK3 | 100 | 97.872 | ENSCPOG00000031679 | DAPK3 | 100 | 94.053 | Cavia_porcellus |
| ENSG00000167657 | DAPK3 | 100 | 98.582 | ENSCLAG00000009010 | DAPK3 | 100 | 95.374 | Chinchilla_lanigera |
| ENSG00000167657 | DAPK3 | 100 | 100.000 | ENSCSAG00000010873 | DAPK3 | 100 | 98.899 | Chlorocebus_sabaeus |
| ENSG00000167657 | DAPK3 | 100 | 94.937 | ENSCPBG00000011124 | DAPK3 | 100 | 79.386 | Chrysemys_picta_bellii |
| ENSG00000167657 | DAPK3 | 100 | 100.000 | ENSCANG00000003009 | DAPK3 | 100 | 98.899 | Colobus_angolensis_palliatus |
| ENSG00000167657 | DAPK3 | 100 | 92.405 | ENSCVAG00000018536 | dapk3 | 98 | 81.124 | Cyprinodon_variegatus |
| ENSG00000167657 | DAPK3 | 100 | 91.139 | ENSDARG00000074447 | dapk3 | 98 | 82.207 | Danio_rerio |
| ENSG00000167657 | DAPK3 | 100 | 100.000 | ENSEASG00005022305 | DAPK3 | 100 | 93.833 | Equus_asinus_asinus |
| ENSG00000167657 | DAPK3 | 100 | 95.732 | ENSECAG00000012570 | DAPK3 | 100 | 93.478 | Equus_caballus |
| ENSG00000167657 | DAPK3 | 100 | 99.367 | ENSFCAG00000013501 | DAPK3 | 100 | 94.053 | Felis_catus |
| ENSG00000167657 | DAPK3 | 100 | 92.405 | ENSFHEG00000009227 | dapk3 | 98 | 81.348 | Fundulus_heteroclitus |
| ENSG00000167657 | DAPK3 | 100 | 91.772 | ENSGMOG00000012530 | dapk3 | 98 | 82.883 | Gadus_morhua |
| ENSG00000167657 | DAPK3 | 100 | 92.405 | ENSGAFG00000020067 | dapk3 | 98 | 81.124 | Gambusia_affinis |
| ENSG00000167657 | DAPK3 | 100 | 89.873 | ENSGACG00000016942 | dapk3 | 98 | 80.674 | Gasterosteus_aculeatus |
| ENSG00000167657 | DAPK3 | 100 | 100.000 | ENSGGOG00000002018 | DAPK3 | 100 | 99.559 | Gorilla_gorilla |
| ENSG00000167657 | DAPK3 | 95 | 93.533 | ENSHGLG00000009769 | DAPK3 | 100 | 93.533 | Heterocephalus_glaber_female |
| ENSG00000167657 | DAPK3 | 100 | 97.163 | ENSHGLG00100012309 | DAPK3 | 100 | 94.934 | Heterocephalus_glaber_male |
| ENSG00000167657 | DAPK3 | 100 | 97.163 | ENSSTOG00000005038 | DAPK3 | 100 | 94.493 | Ictidomys_tridecemlineatus |
| ENSG00000167657 | DAPK3 | 100 | 92.405 | ENSKMAG00000022041 | dapk3 | 98 | 81.757 | Kryptolebias_marmoratus |
| ENSG00000167657 | DAPK3 | 100 | 91.139 | ENSLBEG00000013792 | dapk3 | 99 | 80.580 | Labrus_bergylta |
| ENSG00000167657 | DAPK3 | 100 | 93.038 | ENSLOCG00000001875 | dapk3 | 97 | 83.519 | Lepisosteus_oculatus |
| ENSG00000167657 | DAPK3 | 100 | 100.000 | ENSLAFG00000029621 | DAPK3 | 100 | 92.070 | Loxodonta_africana |
| ENSG00000167657 | DAPK3 | 100 | 100.000 | ENSMFAG00000028991 | DAPK3 | 100 | 98.899 | Macaca_fascicularis |
| ENSG00000167657 | DAPK3 | 100 | 100.000 | ENSMMUG00000022765 | DAPK3 | 100 | 98.899 | Macaca_mulatta |
| ENSG00000167657 | DAPK3 | 100 | 100.000 | ENSMICG00000046071 | DAPK3 | 100 | 97.357 | Microcebus_murinus |
| ENSG00000167657 | DAPK3 | 100 | 91.139 | ENSMMOG00000020516 | dapk3 | 98 | 79.279 | Mola_mola |
| ENSG00000167657 | DAPK3 | 100 | 96.203 | ENSMODG00000000718 | DAPK3 | 100 | 90.529 | Monodelphis_domestica |
| ENSG00000167657 | DAPK3 | 100 | 96.454 | ENSMUSG00000034974 | Dapk3 | 100 | 95.312 | Mus_musculus |
| ENSG00000167657 | DAPK3 | 100 | 99.367 | ENSMPUG00000004467 | DAPK3 | 100 | 94.934 | Mustela_putorius_furo |
| ENSG00000167657 | DAPK3 | 100 | 100.000 | ENSMLUG00000006239 | DAPK3 | 100 | 94.934 | Myotis_lucifugus |
| ENSG00000167657 | DAPK3 | 100 | 96.454 | ENSNGAG00000020954 | Dapk3 | 100 | 83.921 | Nannospalax_galili |
| ENSG00000167657 | DAPK3 | 100 | 96.203 | ENSMEUG00000003969 | DAPK3 | 100 | 87.665 | Notamacropus_eugenii |
| ENSG00000167657 | DAPK3 | 100 | 97.872 | ENSODEG00000014160 | DAPK3 | 100 | 93.612 | Octodon_degus |
| ENSG00000167657 | DAPK3 | 96 | 89.631 | ENSOANG00000003319 | DAPK3 | 100 | 89.631 | Ornithorhynchus_anatinus |
| ENSG00000167657 | DAPK3 | 100 | 91.772 | ENSOMEG00000012168 | dapk3 | 98 | 81.348 | Oryzias_melastigma |
| ENSG00000167657 | DAPK3 | 100 | 99.367 | ENSOGAG00000010293 | DAPK3 | 100 | 96.256 | Otolemur_garnettii |
| ENSG00000167657 | DAPK3 | 100 | 99.367 | ENSOARG00000011263 | DAPK3 | 100 | 92.070 | Ovis_aries |
| ENSG00000167657 | DAPK3 | 100 | 99.780 | ENSPPAG00000029008 | DAPK3 | 100 | 99.780 | Pan_paniscus |
| ENSG00000167657 | DAPK3 | 100 | 98.734 | ENSPPRG00000002025 | DAPK3 | 100 | 89.035 | Panthera_pardus |
| ENSG00000167657 | DAPK3 | 100 | 100.000 | ENSPTRG00000010286 | DAPK3 | 100 | 100.000 | Pan_troglodytes |
| ENSG00000167657 | DAPK3 | 100 | 100.000 | ENSPANG00000018995 | DAPK3 | 100 | 98.899 | Papio_anubis |
| ENSG00000167657 | DAPK3 | 100 | 89.241 | ENSPMGG00000014858 | dapk3 | 95 | 89.189 | Periophthalmus_magnuspinnatus |
| ENSG00000167657 | DAPK3 | 100 | 96.203 | ENSPCIG00000025399 | DAPK3 | 100 | 90.749 | Phascolarctos_cinereus |
| ENSG00000167657 | DAPK3 | 100 | 99.291 | ENSPCAG00000014006 | DAPK3 | 100 | 92.731 | Procavia_capensis |
| ENSG00000167657 | DAPK3 | 100 | 100.000 | ENSPCOG00000023275 | DAPK3 | 100 | 97.577 | Propithecus_coquereli |
| ENSG00000167657 | DAPK3 | 100 | 100.000 | ENSRROG00000045681 | DAPK3 | 100 | 98.899 | Rhinopithecus_roxellana |
| ENSG00000167657 | DAPK3 | 100 | 100.000 | ENSSBOG00000032686 | DAPK3 | 100 | 87.665 | Saimiri_boliviensis_boliviensis |
| ENSG00000167657 | DAPK3 | 100 | 91.139 | ENSSMAG00000015643 | dapk3 | 98 | 80.449 | Scophthalmus_maximus |
| ENSG00000167657 | DAPK3 | 100 | 91.139 | ENSSDUG00000012176 | dapk3 | 99 | 80.088 | Seriola_dumerili |
| ENSG00000167657 | DAPK3 | 100 | 91.139 | ENSSLDG00000011269 | dapk3 | 99 | 79.956 | Seriola_lalandi_dorsalis |
| ENSG00000167657 | DAPK3 | 100 | 91.139 | ENSSPAG00000012830 | dapk3 | 98 | 81.124 | Stegastes_partitus |
| ENSG00000167657 | DAPK3 | 100 | 99.367 | ENSSSCG00000032170 | DAPK3 | 100 | 94.493 | Sus_scrofa |
| ENSG00000167657 | DAPK3 | 100 | 89.873 | ENSTRUG00000006459 | dapk3 | 98 | 79.326 | Takifugu_rubripes |
| ENSG00000167657 | DAPK3 | 100 | 92.405 | ENSTNIG00000014909 | dapk3 | 98 | 81.124 | Tetraodon_nigroviridis |
| ENSG00000167657 | DAPK3 | 100 | 100.000 | ENSTTRG00000006774 | DAPK3 | 100 | 94.053 | Tursiops_truncatus |
| ENSG00000167657 | DAPK3 | 100 | 99.367 | ENSUAMG00000023932 | DAPK3 | 100 | 95.154 | Ursus_americanus |
| ENSG00000167657 | DAPK3 | 100 | 99.367 | ENSVVUG00000004576 | DAPK3 | 100 | 95.374 | Vulpes_vulpes |
| ENSG00000167657 | DAPK3 | 98 | 86.957 | ENSXETG00000006396 | dapk3 | 99 | 79.241 | Xenopus_tropicalis |
| ENSG00000167657 | DAPK3 | 100 | 91.772 | ENSXMAG00000000503 | dapk3 | 98 | 80.899 | Xiphophorus_maculatus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0004674 | protein serine/threonine kinase activity | 21873635. | IBA | Function |
| GO:0004674 | protein serine/threonine kinase activity | 10356987.18239682. | IDA | Function |
| GO:0004674 | protein serine/threonine kinase activity | 16756490. | TAS | Function |
| GO:0005515 | protein binding | 15292222.15910542.21454679.21988832. | IPI | Function |
| GO:0005524 | ATP binding | 21873635. | IBA | Function |
| GO:0005524 | ATP binding | 10356987. | IDA | Function |
| GO:0005634 | nucleus | 21873635. | IBA | Component |
| GO:0005634 | nucleus | - | IDA | Component |
| GO:0005634 | nucleus | - | ISS | Component |
| GO:0005737 | cytoplasm | 21873635. | IBA | Component |
| GO:0005884 | actin filament | - | IEA | Component |
| GO:0006325 | chromatin organization | - | IEA | Process |
| GO:0006355 | regulation of transcription, DNA-templated | - | TAS | Process |
| GO:0006468 | protein phosphorylation | 10356987. | IDA | Process |
| GO:0006915 | apoptotic process | 10580117. | IDA | Process |
| GO:0006915 | apoptotic process | 12917339. | IMP | Process |
| GO:0006940 | regulation of smooth muscle contraction | - | TAS | Process |
| GO:0007088 | regulation of mitotic nuclear division | - | TAS | Process |
| GO:0007346 | regulation of mitotic cell cycle | 21487036. | IMP | Process |
| GO:0008022 | protein C-terminus binding | - | IEA | Function |
| GO:0008140 | cAMP response element binding protein binding | 14685163. | TAS | Function |
| GO:0008360 | regulation of cell shape | - | IEA | Process |
| GO:0010506 | regulation of autophagy | 16756490. | TAS | Process |
| GO:0016605 | PML body | - | IEA | Component |
| GO:0017048 | Rho GTPase binding | 23454120. | IDA | Function |
| GO:0017148 | negative regulation of translation | 18995835. | IDA | Process |
| GO:0030182 | neuron differentiation | - | IEA | Process |
| GO:0030335 | positive regulation of cell migration | - | IEA | Process |
| GO:0035556 | intracellular signal transduction | 10356987. | IDA | Process |
| GO:0042802 | identical protein binding | 18239682.21487036. | IPI | Function |
| GO:0042803 | protein homodimerization activity | 18239682. | IDA | Function |
| GO:0042981 | regulation of apoptotic process | 16756490. | TAS | Process |
| GO:0043065 | positive regulation of apoptotic process | 21487036. | IDA | Process |
| GO:0043519 | regulation of myosin II filament organization | - | IEA | Process |
| GO:0043522 | leucine zipper domain binding | 10580117. | IPI | Function |
| GO:0045121 | membrane raft | - | IEA | Component |
| GO:0046777 | protein autophosphorylation | 18239682. | IDA | Process |
| GO:0046777 | protein autophosphorylation | 16756490. | TAS | Process |
| GO:0051893 | regulation of focal adhesion assembly | 23454120. | IDA | Process |
| GO:0071346 | cellular response to interferon-gamma | 18995835. | IDA | Process |
| GO:0090263 | positive regulation of canonical Wnt signaling pathway | 21454679. | IMP | Process |
| GO:0097190 | apoptotic signaling pathway | - | IEA | Process |
| GO:2000145 | regulation of cell motility | - | TAS | Process |
| GO:2000249 | regulation of actin cytoskeleton reorganization | 23454120. | IDA | Process |
| GO:2000249 | regulation of actin cytoskeleton reorganization | - | TAS | Process |
| GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand | - | IEA | Process |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PRAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| THCA | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| KIRP | |||
| LIHC | |||
| PAAD | |||
| READ | |||
| STAD | |||
| THCA | |||
| UCEC | |||
| UCS |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| SARC | ||
| MESO | ||
| ACC | ||
| SKCM | ||
| PRAD | ||
| ESCA | ||
| COAD | ||
| PAAD | ||
| LAML | ||
| UCEC | ||
| LIHC | ||
| LGG | ||
| UVM |