
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000303276 | RnaseA | PF00074.20 | 6.2e-35 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000304625 | RNASE2-201 | 755 | - | ENSP00000303276 | 161 (aa) | - | P10153 |
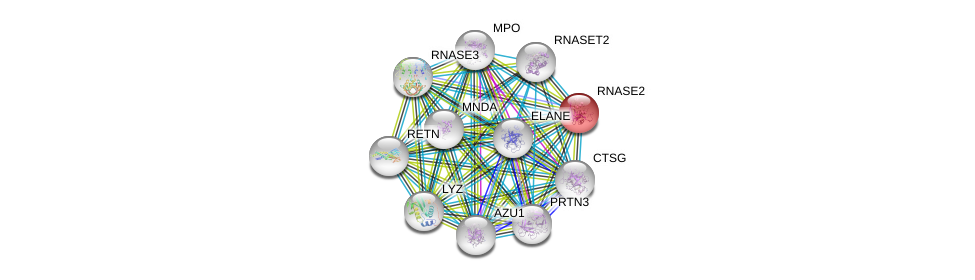
| Ensembl ID | Gene Symbol | Coverage | Identiy | Paralog | Gene Symbol | Coverage | Identiy |
|---|---|---|---|---|---|---|---|
| ENSG00000169385 | RNASE2 | 88 | 41.844 | ENSG00000173431 | RNASE8 | 87 | 41.844 |
| ENSG00000169385 | RNASE2 | 85 | 45.255 | ENSG00000169413 | RNASE6 | 87 | 45.255 |
| ENSG00000169385 | RNASE2 | 88 | 39.716 | ENSG00000165799 | RNASE7 | 87 | 39.716 |
| ENSG00000169385 | RNASE2 | 77 | 33.077 | ENSG00000129538 | RNASE1 | 89 | 33.621 |
| ENSG00000169385 | RNASE2 | 100 | 68.944 | ENSG00000169397 | RNASE3 | 100 | 68.944 |
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000169385 | RNASE2 | 100 | 73.292 | ENSANAG00000014479 | - | 100 | 73.292 | Aotus_nancymaae |
| ENSG00000169385 | RNASE2 | 100 | 54.658 | ENSBTAG00000049914 | - | 86 | 54.658 | Bos_taurus |
| ENSG00000169385 | RNASE2 | 86 | 57.971 | ENSBTAG00000025664 | - | 86 | 56.522 | Bos_taurus |
| ENSG00000169385 | RNASE2 | 100 | 70.186 | ENSCJAG00000009638 | - | 100 | 70.186 | Callithrix_jacchus |
| ENSG00000169385 | RNASE2 | 86 | 53.623 | ENSCHIG00000006998 | - | 100 | 53.416 | Capra_hircus |
| ENSG00000169385 | RNASE2 | 100 | 71.429 | ENSCCAG00000008336 | - | 100 | 71.429 | Cebus_capucinus |
| ENSG00000169385 | RNASE2 | 100 | 65.839 | ENSCATG00000021993 | RNASE3 | 100 | 65.839 | Cercocebus_atys |
| ENSG00000169385 | RNASE2 | 100 | 83.230 | ENSCSAG00000000344 | RNASE2 | 100 | 83.230 | Chlorocebus_sabaeus |
| ENSG00000169385 | RNASE2 | 99 | 74.375 | ENSCANG00000041784 | RNASE2 | 99 | 74.375 | Colobus_angolensis_palliatus |
| ENSG00000169385 | RNASE2 | 89 | 48.276 | ENSCGRG00001015465 | Ear11 | 99 | 49.689 | Cricetulus_griseus_chok1gshd |
| ENSG00000169385 | RNASE2 | 90 | 54.110 | ENSCGRG00001015384 | Ear7 | 100 | 54.938 | Cricetulus_griseus_chok1gshd |
| ENSG00000169385 | RNASE2 | 55 | 48.864 | ENSCGRG00001009946 | - | 71 | 47.009 | Cricetulus_griseus_chok1gshd |
| ENSG00000169385 | RNASE2 | 99 | 49.068 | ENSCGRG00001015041 | Ear10 | 99 | 49.068 | Cricetulus_griseus_chok1gshd |
| ENSG00000169385 | RNASE2 | 55 | 48.864 | ENSCGRG00000013872 | - | 71 | 47.009 | Cricetulus_griseus_crigri |
| ENSG00000169385 | RNASE2 | 89 | 48.276 | ENSCGRG00000013443 | Ear10 | 99 | 49.689 | Cricetulus_griseus_crigri |
| ENSG00000169385 | RNASE2 | 90 | 54.110 | ENSCGRG00000006180 | Ear7 | 100 | 54.938 | Cricetulus_griseus_crigri |
| ENSG00000169385 | RNASE2 | 89 | 48.276 | ENSCGRG00000003290 | Ear11 | 99 | 49.689 | Cricetulus_griseus_crigri |
| ENSG00000169385 | RNASE2 | 99 | 55.000 | ENSDNOG00000033465 | - | 99 | 55.000 | Dasypus_novemcinctus |
| ENSG00000169385 | RNASE2 | 99 | 48.125 | ENSDNOG00000043774 | - | 99 | 48.125 | Dasypus_novemcinctus |
| ENSG00000169385 | RNASE2 | 86 | 49.275 | ENSDORG00000028426 | - | 84 | 50.000 | Dipodomys_ordii |
| ENSG00000169385 | RNASE2 | 85 | 54.348 | ENSEASG00005010211 | - | 85 | 54.545 | Equus_asinus_asinus |
| ENSG00000169385 | RNASE2 | 99 | 54.037 | ENSECAG00000002332 | - | 99 | 54.037 | Equus_caballus |
| ENSG00000169385 | RNASE2 | 100 | 99.379 | ENSGGOG00000043975 | RNASE2 | 100 | 99.379 | Gorilla_gorilla |
| ENSG00000169385 | RNASE2 | 100 | 68.944 | ENSGGOG00000037139 | RNASE3 | 100 | 68.944 | Gorilla_gorilla |
| ENSG00000169385 | RNASE2 | 88 | 47.887 | ENSSTOG00000030965 | - | 87 | 46.203 | Ictidomys_tridecemlineatus |
| ENSG00000169385 | RNASE2 | 99 | 50.000 | ENSSTOG00000020133 | - | 99 | 50.000 | Ictidomys_tridecemlineatus |
| ENSG00000169385 | RNASE2 | 98 | 50.633 | ENSSTOG00000024645 | - | 98 | 50.633 | Ictidomys_tridecemlineatus |
| ENSG00000169385 | RNASE2 | 100 | 68.944 | ENSMFAG00000014172 | - | 100 | 68.944 | Macaca_fascicularis |
| ENSG00000169385 | RNASE2 | 100 | 65.217 | ENSMFAG00000014110 | - | 100 | 65.217 | Macaca_fascicularis |
| ENSG00000169385 | RNASE2 | 100 | 68.944 | ENSMMUG00000037666 | - | 100 | 68.944 | Macaca_mulatta |
| ENSG00000169385 | RNASE2 | 100 | 81.988 | ENSMMUG00000046252 | RNASE2 | 100 | 81.988 | Macaca_mulatta |
| ENSG00000169385 | RNASE2 | 100 | 66.460 | ENSMMUG00000040314 | - | 100 | 66.460 | Macaca_mulatta |
| ENSG00000169385 | RNASE2 | 100 | 68.944 | ENSMNEG00000001455 | - | 100 | 68.944 | Macaca_nemestrina |
| ENSG00000169385 | RNASE2 | 100 | 67.081 | ENSMNEG00000038266 | - | 100 | 67.081 | Macaca_nemestrina |
| ENSG00000169385 | RNASE2 | 100 | 64.596 | ENSMLEG00000002481 | RNASE3 | 100 | 64.596 | Mandrillus_leucophaeus |
| ENSG00000169385 | RNASE2 | 83 | 55.970 | ENSMAUG00000016887 | - | 83 | 55.970 | Mesocricetus_auratus |
| ENSG00000169385 | RNASE2 | 80 | 49.231 | ENSMUSG00000059606 | Rnase2b | 82 | 49.231 | Mus_musculus |
| ENSG00000169385 | RNASE2 | 77 | 53.600 | ENSMUSG00000050766 | Ear14 | 80 | 53.175 | Mus_musculus |
| ENSG00000169385 | RNASE2 | 81 | 49.618 | ENSMUSG00000047222 | Rnase2a | 83 | 49.618 | Mus_musculus |
| ENSG00000169385 | RNASE2 | 98 | 47.799 | ENSMUSG00000090166 | Ear10 | 98 | 47.799 | Mus_musculus |
| ENSG00000169385 | RNASE2 | 78 | 51.969 | ENSMUSG00000062148 | Ear6 | 98 | 51.572 | Mus_musculus |
| ENSG00000169385 | RNASE2 | 78 | 48.819 | ENSMUSG00000072596 | Ear2 | 98 | 47.799 | Mus_musculus |
| ENSG00000169385 | RNASE2 | 78 | 49.606 | ENSMUSG00000072601 | Ear1 | 98 | 48.428 | Mus_musculus |
| ENSG00000169385 | RNASE2 | 65 | 51.429 | MGP_PahariEiJ_G0030246 | - | 97 | 51.429 | Mus_pahari |
| ENSG00000169385 | RNASE2 | 87 | 51.429 | ENSMLUG00000023348 | - | 100 | 51.429 | Myotis_lucifugus |
| ENSG00000169385 | RNASE2 | 100 | 48.193 | ENSMLUG00000030121 | - | 100 | 48.193 | Myotis_lucifugus |
| ENSG00000169385 | RNASE2 | 86 | 51.449 | ENSOCUG00000007497 | - | 99 | 49.682 | Oryctolagus_cuniculus |
| ENSG00000169385 | RNASE2 | 100 | 44.172 | ENSOCUG00000015951 | - | 100 | 44.172 | Oryctolagus_cuniculus |
| ENSG00000169385 | RNASE2 | 99 | 61.250 | ENSOGAG00000013322 | - | 100 | 61.250 | Otolemur_garnettii |
| ENSG00000169385 | RNASE2 | 100 | 54.658 | ENSOGAG00000032016 | - | 100 | 54.658 | Otolemur_garnettii |
| ENSG00000169385 | RNASE2 | 100 | 59.006 | ENSOGAG00000027626 | - | 100 | 59.006 | Otolemur_garnettii |
| ENSG00000169385 | RNASE2 | 100 | 53.416 | ENSOARG00000019719 | - | 100 | 53.416 | Ovis_aries |
| ENSG00000169385 | RNASE2 | 100 | 98.758 | ENSPPAG00000033117 | RNASE2 | 100 | 98.758 | Pan_paniscus |
| ENSG00000169385 | RNASE2 | 100 | 99.379 | ENSPTRG00000006106 | RNASE2 | 100 | 99.379 | Pan_troglodytes |
| ENSG00000169385 | RNASE2 | 100 | 68.323 | ENSPANG00000005471 | - | 100 | 68.323 | Papio_anubis |
| ENSG00000169385 | RNASE2 | 89 | 52.448 | ENSPEMG00000023630 | - | 100 | 54.037 | Peromyscus_maniculatus_bairdii |
| ENSG00000169385 | RNASE2 | 100 | 77.019 | ENSPPYG00000005579 | - | 100 | 77.019 | Pongo_abelii |
| ENSG00000169385 | RNASE2 | 100 | 93.168 | ENSPPYG00000005580 | RNASE2 | 100 | 93.168 | Pongo_abelii |
| ENSG00000169385 | RNASE2 | 83 | 51.111 | ENSRNOG00000031445 | LOC100361866 | 100 | 50.000 | Rattus_norvegicus |
| ENSG00000169385 | RNASE2 | 80 | 52.308 | ENSRNOG00000032133 | Rnase2 | 81 | 52.344 | Rattus_norvegicus |
| ENSG00000169385 | RNASE2 | 80 | 53.846 | ENSRNOG00000025515 | Ear1 | 100 | 49.383 | Rattus_norvegicus |
| ENSG00000169385 | RNASE2 | 83 | 51.111 | ENSRNOG00000050295 | Rnase17 | 100 | 50.617 | Rattus_norvegicus |
| ENSG00000169385 | RNASE2 | 83 | 51.111 | ENSRNOG00000053457 | Rnase3 | 100 | 50.000 | Rattus_norvegicus |
| ENSG00000169385 | RNASE2 | 100 | 65.217 | ENSRBIG00000010077 | RNASE3 | 100 | 65.217 | Rhinopithecus_bieti |
| ENSG00000169385 | RNASE2 | 100 | 73.913 | ENSSBOG00000007632 | - | 100 | 73.913 | Saimiri_boliviensis_boliviensis |
| ENSG00000169385 | RNASE2 | 86 | 52.899 | ENSSSCG00000035377 | - | 89 | 52.113 | Sus_scrofa |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0001530 | lipopolysaccharide binding | 23992292. | IMP | Function |
| GO:0003676 | nucleic acid binding | - | IEA | Function |
| GO:0004522 | ribonuclease A activity | - | IEA | Function |
| GO:0004540 | ribonuclease activity | 21873635. | IBA | Function |
| GO:0004540 | ribonuclease activity | 9826755. | IDA | Function |
| GO:0005576 | extracellular region | - | TAS | Component |
| GO:0006401 | RNA catabolic process | 2734298. | TAS | Process |
| GO:0006935 | chemotaxis | 12855582. | IDA | Process |
| GO:0035578 | azurophil granule lumen | - | TAS | Component |
| GO:0043152 | induction of bacterial agglutination | 23992292. | IMP | Process |
| GO:0043312 | neutrophil degranulation | - | TAS | Process |
| GO:0050829 | defense response to Gram-negative bacterium | 23992292. | IMP | Process |
| GO:0050830 | defense response to Gram-positive bacterium | 23992292. | IMP | Process |
| GO:0051607 | defense response to virus | 9826755. | IDA | Process |
| GO:0070062 | extracellular exosome | 23533145. | HDA | Component |
| GO:0090501 | RNA phosphodiester bond hydrolysis | 9826755. | IDA | Process |
| GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic | - | IEA | Process |
| PID | Title | Article | Time | Author | Doi |
|---|---|---|---|---|---|
| 16428483 |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| COAD | |||||||
| DLBC | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| LAML | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| ACC | |||
| ACC | |||
| CHOL | |||
| HNSC | |||
| KIRP | |||
| LUSC | |||
| OV | |||
| PAAD | |||
| READ |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| STAD | ||
| SKCM | ||
| PAAD | ||
| PCPG | ||
| GBM | ||
| LIHC | ||
| LGG |