
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000477548 | DFFB-209 | 2923 | - | ENSP00000462175 | 103 (aa) | - | G3XAD0 |
| ENST00000378206 | DFFB-204 | 2842 | - | ENSP00000367448 | 80 (aa) | - | Q96P72 |
| ENST00000491998 | DFFB-211 | 3152 | - | ENSP00000436775 | 270 (aa) | - | O76075 |
| ENST00000468793 | DFFB-207 | 2986 | - | ENSP00000463688 | 115 (aa) | - | Q96P73 |
| ENST00000378209 | DFFB-205 | 3028 | XM_017000498 | ENSP00000367454 | 338 (aa) | XP_016855987 | O76075 |
| ENST00000475969 | DFFB-208 | 2731 | - | ENSP00000463704 | 115 (aa) | - | Q96P73 |
| ENST00000339350 | DFFB-202 | 3005 | - | ENSP00000343218 | 116 (aa) | - | O76075 |
| ENST00000461150 | DFFB-206 | 686 | - | ENSP00000462770 | 78 (aa) | - | J3KT26 |
| ENST00000338895 | DFFB-201 | 3105 | XM_017000499 | ENSP00000339524 | 362 (aa) | XP_016855988 | B4DZS0 |
| ENST00000481945 | DFFB-210 | 636 | - | - | - (aa) | - | - |
| ENST00000625756 | DFFB-212 | 649 | - | ENSP00000486916 | 103 (aa) | - | G3XAD0 |
| ENST00000341385 | DFFB-203 | 572 | - | ENSP00000345906 | 82 (aa) | - | A0A0B4J1S5 |
| Pathway ID | Pathway Name | Source |
|---|---|---|
| hsa04210 | Apoptosis | KEGG |
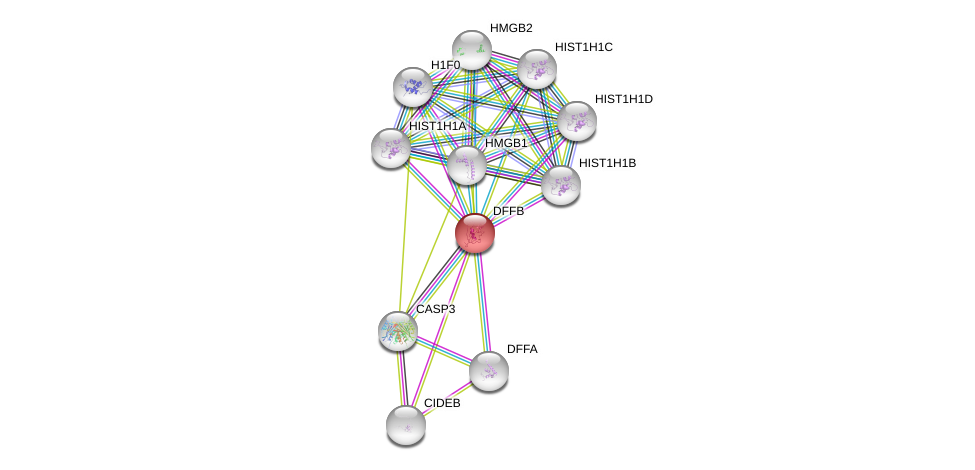
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000169598 | DFFB | 100 | 77.219 | ENSMUSG00000029027 | Dffb | 98 | 77.219 | Mus_musculus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000790 | nuclear chromatin | 21873635. | IBA | Component |
| GO:0000790 | nuclear chromatin | 19882353. | IDA | Component |
| GO:0000790 | nuclear chromatin | 22253444. | IDA | Component |
| GO:0004536 | deoxyribonuclease activity | 21873635. | IBA | Function |
| GO:0004536 | deoxyribonuclease activity | 19944011. | IMP | Function |
| GO:0005515 | protein binding | 19882353. | IPI | Function |
| GO:0005634 | nucleus | 10959840.15572351.22253444. | IDA | Component |
| GO:0005654 | nucleoplasm | - | TAS | Component |
| GO:0005730 | nucleolus | - | IDA | Component |
| GO:0005829 | cytosol | 9108473.22253444. | IDA | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0006308 | DNA catabolic process | 19944011. | IMP | Process |
| GO:0006309 | apoptotic DNA fragmentation | 21873635. | IBA | Process |
| GO:0006309 | apoptotic DNA fragmentation | 15572351.22253444. | IDA | Process |
| GO:0006309 | apoptotic DNA fragmentation | - | TAS | Process |
| GO:0019899 | enzyme binding | 10959840. | IPI | Function |
| GO:0019904 | protein domain specific binding | 19944011. | IPI | Function |
| GO:0030263 | apoptotic chromosome condensation | 10959840. | IDA | Process |
| GO:0042802 | identical protein binding | 11371636. | IMP | Function |
| GO:0051260 | protein homooligomerization | 11371636. | IMP | Process |
| GO:0097718 | disordered domain specific binding | 11371636. | IPI | Function |