
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000307630 | YWHAG-201 | 3750 | - | ENSP00000306330 | 247 (aa) | - | P61981 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000170027 | rs17149161 | 7 | 76348912 | A | Multiple sclerosis | 21654844 | [NR] | 1.87 | EFO_0003885 |
| ENSG00000170027 | rs7779014 | 7 | 76346269 | T | Multiple sclerosis | 21654844 | [NR] unit increase | 0.48 | EFO_0003885 |
| ENSG00000170027 | rs2961033 | 7 | 76335026 | C | Schizophrenia | 30285260 | [1.05-1.12] | 1.082 | EFO_0000692 |
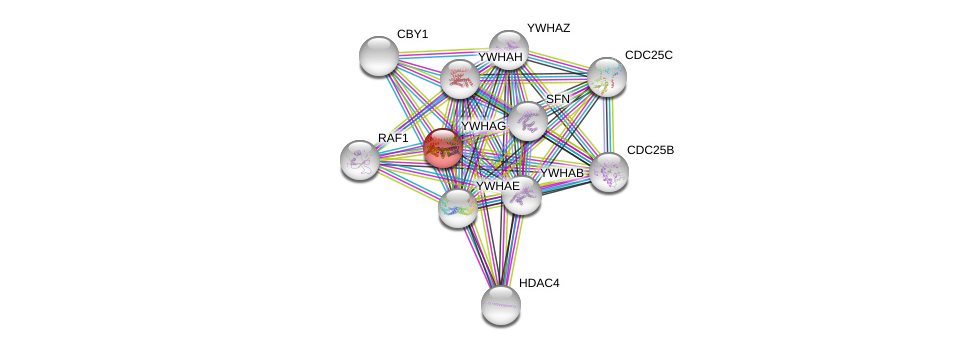
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000170027 | YWHAG | 99 | 86.992 | ENSG00000128245 | YWHAH | 100 | 86.992 | Homo_sapiens |
| ENSG00000170027 | YWHAG | 100 | 63.968 | ENSG00000108953 | YWHAE | 99 | 71.930 | Homo_sapiens |
| ENSG00000170027 | YWHAG | 100 | 100.000 | ENSMUSG00000051391 | Ywhag | 100 | 100.000 | Mus_musculus |
| ENSG00000170027 | YWHAG | 99 | 86.992 | ENSMUSG00000018965 | Ywhah | 100 | 86.992 | Mus_musculus |
| ENSG00000170027 | YWHAG | 100 | 63.968 | ENSMUSG00000020849 | Ywhae | 95 | 63.968 | Mus_musculus |
| ENSG00000170027 | YWHAG | 94 | 65.385 | YDR099W | BMH2 | 85 | 65.385 | Saccharomyces_cerevisiae |
| ENSG00000170027 | YWHAG | 98 | 62.810 | YER177W | BMH1 | 90 | 62.810 | Saccharomyces_cerevisiae |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000086 | G2/M transition of mitotic cell cycle | - | TAS | Process |
| GO:0003723 | RNA binding | 22658674. | HDA | Function |
| GO:0005080 | protein kinase C binding | 10433554. | IPI | Function |
| GO:0005159 | insulin-like growth factor receptor binding | 12482592. | IPI | Function |
| GO:0005515 | protein binding | 11237865.11504882.12364343.15696159.16511560.16511572.16516142.16959763.17085597.17159145.18045992.18458160.18812399.19172738.19640509.19860830.19933256.20639859.20642453.20936779.21044950.21988832.22653443.23075850.23572552.23622247.23752268.24255178.24947832.25416956.25852190.26638075.28330616.30021884. | IPI | Function |
| GO:0005739 | mitochondrion | - | IEA | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0005925 | focal adhesion | 21423176. | HDA | Component |
| GO:0006469 | negative regulation of protein kinase activity | 10433554. | NAS | Process |
| GO:0006605 | protein targeting | - | IEA | Process |
| GO:0008426 | protein kinase C inhibitor activity | 10433554. | NAS | Function |
| GO:0009966 | regulation of signal transduction | 10433554.10486217. | NAS | Process |
| GO:0010389 | regulation of G2/M transition of mitotic cell cycle | - | TAS | Process |
| GO:0016020 | membrane | 19946888. | HDA | Component |
| GO:0019904 | protein domain specific binding | - | IEA | Function |
| GO:0030971 | receptor tyrosine kinase binding | - | IEA | Function |
| GO:0032869 | cellular response to insulin stimulus | - | IEA | Process |
| GO:0042802 | identical protein binding | 17085597.20639859. | IPI | Function |
| GO:0045664 | regulation of neuron differentiation | 11824616. | IMP | Process |
| GO:0048167 | regulation of synaptic plasticity | 11824616. | IMP | Process |
| GO:0061024 | membrane organization | - | TAS | Process |
| GO:0070062 | extracellular exosome | 19056867.19199708.20458337. | HDA | Component |
| GO:0071901 | negative regulation of protein serine/threonine kinase activity | - | IEA | Process |
| GO:0097711 | ciliary basal body-plasma membrane docking | - | TAS | Process |
| GO:0098793 | presynapse | - | IEA | Component |
| GO:1900740 | positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway | - | TAS | Process |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRP | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| SARC | |||||||
| SARC | |||||||
| SARC | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| DLBC | |||
| TGCT | |||
| THCA |
| Cancer | P-value | Q-value |
|---|---|---|
| MESO | ||
| HNSC | ||
| PRAD | ||
| BRCA | ||
| KIRP | ||
| COAD | ||
| PAAD | ||
| CESC | ||
| LAML | ||
| KICH | ||
| UCEC | ||
| GBM | ||
| LIHC | ||
| LUAD | ||
| UVM |