
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000435940 | zf-C2H2 | PF00096.26 | 1.2e-33 | 1 | 5 |
| ENSP00000435940 | zf-C2H2 | PF00096.26 | 1.2e-33 | 2 | 5 |
| ENSP00000435940 | zf-C2H2 | PF00096.26 | 1.2e-33 | 3 | 5 |
| ENSP00000435940 | zf-C2H2 | PF00096.26 | 1.2e-33 | 4 | 5 |
| ENSP00000435940 | zf-C2H2 | PF00096.26 | 1.2e-33 | 5 | 5 |
| ENSP00000302669 | zf-C2H2 | PF00096.26 | 6.6e-33 | 1 | 5 |
| ENSP00000302669 | zf-C2H2 | PF00096.26 | 6.6e-33 | 2 | 5 |
| ENSP00000302669 | zf-C2H2 | PF00096.26 | 6.6e-33 | 3 | 5 |
| ENSP00000302669 | zf-C2H2 | PF00096.26 | 6.6e-33 | 4 | 5 |
| ENSP00000302669 | zf-C2H2 | PF00096.26 | 6.6e-33 | 5 | 5 |
| ENSP00000398431 | zf-C2H2 | PF00096.26 | 9.9e-33 | 1 | 5 |
| ENSP00000398431 | zf-C2H2 | PF00096.26 | 9.9e-33 | 2 | 5 |
| ENSP00000398431 | zf-C2H2 | PF00096.26 | 9.9e-33 | 3 | 5 |
| ENSP00000398431 | zf-C2H2 | PF00096.26 | 9.9e-33 | 4 | 5 |
| ENSP00000398431 | zf-C2H2 | PF00096.26 | 9.9e-33 | 5 | 5 |
| ENSP00000432237 | zf-C2H2 | PF00096.26 | 1.1e-32 | 1 | 5 |
| ENSP00000432237 | zf-C2H2 | PF00096.26 | 1.1e-32 | 2 | 5 |
| ENSP00000432237 | zf-C2H2 | PF00096.26 | 1.1e-32 | 3 | 5 |
| ENSP00000432237 | zf-C2H2 | PF00096.26 | 1.1e-32 | 4 | 5 |
| ENSP00000432237 | zf-C2H2 | PF00096.26 | 1.1e-32 | 5 | 5 |
| ENSP00000431262 | zf-C2H2 | PF00096.26 | 1.8e-32 | 1 | 5 |
| ENSP00000431262 | zf-C2H2 | PF00096.26 | 1.8e-32 | 2 | 5 |
| ENSP00000431262 | zf-C2H2 | PF00096.26 | 1.8e-32 | 3 | 5 |
| ENSP00000431262 | zf-C2H2 | PF00096.26 | 1.8e-32 | 4 | 5 |
| ENSP00000431262 | zf-C2H2 | PF00096.26 | 1.8e-32 | 5 | 5 |
| ENSP00000354118 | zf-C2H2 | PF00096.26 | 2.6e-32 | 1 | 5 |
| ENSP00000354118 | zf-C2H2 | PF00096.26 | 2.6e-32 | 2 | 5 |
| ENSP00000354118 | zf-C2H2 | PF00096.26 | 2.6e-32 | 3 | 5 |
| ENSP00000354118 | zf-C2H2 | PF00096.26 | 2.6e-32 | 4 | 5 |
| ENSP00000354118 | zf-C2H2 | PF00096.26 | 2.6e-32 | 5 | 5 |
| ENSP00000351686 | zf-C2H2 | PF00096.26 | 2.8e-32 | 1 | 5 |
| ENSP00000351686 | zf-C2H2 | PF00096.26 | 2.8e-32 | 2 | 5 |
| ENSP00000351686 | zf-C2H2 | PF00096.26 | 2.8e-32 | 3 | 5 |
| ENSP00000351686 | zf-C2H2 | PF00096.26 | 2.8e-32 | 4 | 5 |
| ENSP00000351686 | zf-C2H2 | PF00096.26 | 2.8e-32 | 5 | 5 |
| ENSP00000435940 | zf-met | PF12874.7 | 2.1e-08 | 1 | 2 |
| ENSP00000435940 | zf-met | PF12874.7 | 2.1e-08 | 2 | 2 |
| ENSP00000302669 | zf-met | PF12874.7 | 2.3e-05 | 1 | 2 |
| ENSP00000302669 | zf-met | PF12874.7 | 2.3e-05 | 2 | 2 |
| ENSP00000398431 | zf-met | PF12874.7 | 2.6e-05 | 1 | 2 |
| ENSP00000398431 | zf-met | PF12874.7 | 2.6e-05 | 2 | 2 |
| ENSP00000432237 | zf-met | PF12874.7 | 2.7e-05 | 1 | 2 |
| ENSP00000432237 | zf-met | PF12874.7 | 2.7e-05 | 2 | 2 |
| ENSP00000431262 | zf-met | PF12874.7 | 3.1e-05 | 1 | 2 |
| ENSP00000431262 | zf-met | PF12874.7 | 3.1e-05 | 2 | 2 |
| ENSP00000354118 | zf-met | PF12874.7 | 3.5e-05 | 1 | 2 |
| ENSP00000354118 | zf-met | PF12874.7 | 3.5e-05 | 2 | 2 |
| ENSP00000351686 | zf-met | PF12874.7 | 3.5e-05 | 1 | 2 |
| ENSP00000351686 | zf-met | PF12874.7 | 3.5e-05 | 2 | 2 |
| ENSP00000435940 | zf-C2H2_jaz | PF12171.8 | 3.7e-19 | 1 | 1 |
| ENSP00000302669 | zf-C2H2_jaz | PF12171.8 | 7.3e-17 | 1 | 1 |
| ENSP00000398431 | zf-C2H2_jaz | PF12171.8 | 1.1e-16 | 1 | 1 |
| ENSP00000432237 | zf-C2H2_jaz | PF12171.8 | 1.2e-16 | 1 | 1 |
| ENSP00000431262 | zf-C2H2_jaz | PF12171.8 | 1.8e-16 | 1 | 1 |
| ENSP00000354118 | zf-C2H2_jaz | PF12171.8 | 2.7e-16 | 1 | 1 |
| ENSP00000351686 | zf-C2H2_jaz | PF12171.8 | 2.8e-16 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000527581 | PRDM10-206 | 620 | - | ENSP00000432093 | 139 (aa) | - | E9PS52 |
| ENST00000531431 | PRDM10-208 | 528 | - | ENSP00000436681 | 115 (aa) | - | E9PIB4 |
| ENST00000304538 | PRDM10-201 | 5817 | - | ENSP00000302669 | 1023 (aa) | - | Q9NQV6 |
| ENST00000533431 | PRDM10-209 | 2643 | - | ENSP00000435940 | 865 (aa) | - | E9PRS0 |
| ENST00000526082 | PRDM10-205 | 3308 | - | ENSP00000432237 | 1074 (aa) | - | Q9NQV6 |
| ENST00000360871 | PRDM10-203 | 6301 | XM_011542912 | ENSP00000354118 | 1156 (aa) | XP_011541214 | Q9NQV6 |
| ENST00000528746 | PRDM10-207 | 6182 | - | ENSP00000431262 | 1117 (aa) | - | E9PLV1 |
| ENST00000358825 | PRDM10-202 | 6322 | - | ENSP00000351686 | 1160 (aa) | - | Q9NQV6 |
| ENST00000423662 | PRDM10-204 | 5998 | XM_017018023 | ENSP00000398431 | 1061 (aa) | XP_016873512 | Q9NQV6 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000170325 | rs145658364 | 11 | 129988672 | ? | DNA methylation variation (age effect) | 30348214 | GO_0006306 | ||
| ENSG00000170325 | rs111703287 | 11 | 129982285 | T | Carotid intima media thickness in rheumatoid arthritis | 30251476 | [0.074-0.164] unit increase | 0.119 | EFO_0000685|EFO_0007117 |
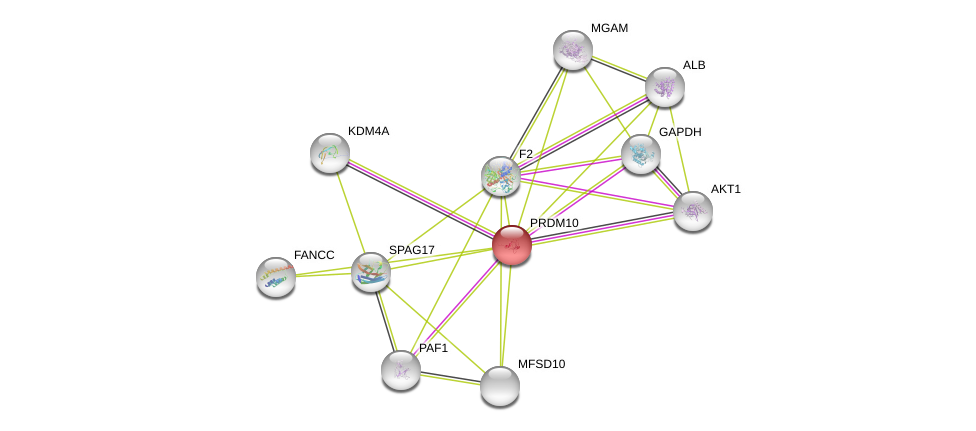
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000170325 | PRDM10 | 100 | 94.104 | ENSAMEG00000002425 | PRDM10 | 100 | 91.402 | Ailuropoda_melanoleuca |
| ENSG00000170325 | PRDM10 | 99 | 86.207 | ENSAPLG00000004858 | PRDM10 | 100 | 85.026 | Anas_platyrhynchos |
| ENSG00000170325 | PRDM10 | 100 | 89.515 | ENSANAG00000022064 | PRDM10 | 100 | 83.491 | Aotus_nancymaae |
| ENSG00000170325 | PRDM10 | 100 | 93.534 | ENSBTAG00000019001 | PRDM10 | 100 | 94.118 | Bos_taurus |
| ENSG00000170325 | PRDM10 | 100 | 98.497 | ENSCJAG00000009235 | PRDM10 | 100 | 98.190 | Callithrix_jacchus |
| ENSG00000170325 | PRDM10 | 100 | 93.873 | ENSCAFG00000010164 | PRDM10 | 100 | 93.253 | Canis_familiaris |
| ENSG00000170325 | PRDM10 | 100 | 94.718 | ENSCAFG00020010152 | PRDM10 | 100 | 94.718 | Canis_lupus_dingo |
| ENSG00000170325 | PRDM10 | 100 | 93.858 | ENSCHIG00000026530 | PRDM10 | 100 | 93.253 | Capra_hircus |
| ENSG00000170325 | PRDM10 | 100 | 98.103 | ENSTSYG00000000544 | PRDM10 | 100 | 98.139 | Carlito_syrichta |
| ENSG00000170325 | PRDM10 | 100 | 85.458 | ENSCAPG00000017296 | PRDM10 | 98 | 87.638 | Cavia_aperea |
| ENSG00000170325 | PRDM10 | 99 | 81.504 | ENSCPOG00000000389 | PRDM10 | 98 | 86.040 | Cavia_porcellus |
| ENSG00000170325 | PRDM10 | 100 | 98.382 | ENSCCAG00000024567 | PRDM10 | 100 | 98.190 | Cebus_capucinus |
| ENSG00000170325 | PRDM10 | 100 | 98.844 | ENSCATG00000040031 | PRDM10 | 100 | 98.789 | Cercocebus_atys |
| ENSG00000170325 | PRDM10 | 100 | 93.037 | ENSCLAG00000014605 | PRDM10 | 99 | 97.403 | Chinchilla_lanigera |
| ENSG00000170325 | PRDM10 | 100 | 98.875 | ENSCSAG00000002202 | PRDM10 | 100 | 98.875 | Chlorocebus_sabaeus |
| ENSG00000170325 | PRDM10 | 99 | 85.977 | ENSCPBG00000026880 | PRDM10 | 99 | 82.255 | Chrysemys_picta_bellii |
| ENSG00000170325 | PRDM10 | 100 | 99.075 | ENSCANG00000032669 | PRDM10 | 100 | 98.879 | Colobus_angolensis_palliatus |
| ENSG00000170325 | PRDM10 | 100 | 94.335 | ENSCGRG00001021647 | Prdm10 | 98 | 92.494 | Cricetulus_griseus_chok1gshd |
| ENSG00000170325 | PRDM10 | 100 | 94.335 | ENSCGRG00000006562 | Prdm10 | 98 | 92.494 | Cricetulus_griseus_crigri |
| ENSG00000170325 | PRDM10 | 86 | 91.348 | ENSDNOG00000001366 | PRDM10 | 95 | 94.292 | Dasypus_novemcinctus |
| ENSG00000170325 | PRDM10 | 100 | 94.798 | ENSDORG00000014292 | Prdm10 | 100 | 93.945 | Dipodomys_ordii |
| ENSG00000170325 | PRDM10 | 100 | 95.242 | ENSEASG00005022914 | PRDM10 | 100 | 95.242 | Equus_asinus_asinus |
| ENSG00000170325 | PRDM10 | 100 | 95.329 | ENSECAG00000011499 | PRDM10 | 100 | 95.156 | Equus_caballus |
| ENSG00000170325 | PRDM10 | 100 | 94.220 | ENSFCAG00000004111 | PRDM10 | 96 | 94.709 | Felis_catus |
| ENSG00000170325 | PRDM10 | 99 | 85.813 | ENSFALG00000004873 | PRDM10 | 100 | 80.667 | Ficedula_albicollis |
| ENSG00000170325 | PRDM10 | 100 | 93.799 | ENSFDAG00000017311 | PRDM10 | 100 | 91.794 | Fukomys_damarensis |
| ENSG00000170325 | PRDM10 | 99 | 86.552 | ENSGALG00000033660 | PRDM10 | 99 | 85.586 | Gallus_gallus |
| ENSG00000170325 | PRDM10 | 98 | 63.877 | ENSGACG00000012293 | prdm10 | 99 | 67.901 | Gasterosteus_aculeatus |
| ENSG00000170325 | PRDM10 | 100 | 83.805 | ENSGAGG00000012523 | PRDM10 | 99 | 83.719 | Gopherus_agassizii |
| ENSG00000170325 | PRDM10 | 100 | 100.000 | ENSGGOG00000015359 | PRDM10 | 100 | 99.397 | Gorilla_gorilla |
| ENSG00000170325 | PRDM10 | 100 | 93.658 | ENSHGLG00000001106 | PRDM10 | 100 | 92.249 | Heterocephalus_glaber_female |
| ENSG00000170325 | PRDM10 | 100 | 94.211 | ENSHGLG00100002350 | PRDM10 | 100 | 92.577 | Heterocephalus_glaber_male |
| ENSG00000170325 | PRDM10 | 100 | 96.416 | ENSSTOG00000002015 | PRDM10 | 100 | 96.021 | Ictidomys_tridecemlineatus |
| ENSG00000170325 | PRDM10 | 93 | 93.433 | ENSJJAG00000021493 | Prdm10 | 100 | 83.275 | Jaculus_jaculus |
| ENSG00000170325 | PRDM10 | 100 | 92.775 | ENSLAFG00000031867 | PRDM10 | 100 | 91.402 | Loxodonta_africana |
| ENSG00000170325 | PRDM10 | 100 | 99.281 | ENSMFAG00000031008 | PRDM10 | 100 | 98.966 | Macaca_fascicularis |
| ENSG00000170325 | PRDM10 | 100 | 99.281 | ENSMMUG00000017555 | PRDM10 | 100 | 98.966 | Macaca_mulatta |
| ENSG00000170325 | PRDM10 | 100 | 99.281 | ENSMNEG00000036493 | PRDM10 | 100 | 98.879 | Macaca_nemestrina |
| ENSG00000170325 | PRDM10 | 94 | 98.824 | ENSMLEG00000042028 | PRDM10 | 100 | 98.824 | Mandrillus_leucophaeus |
| ENSG00000170325 | PRDM10 | 99 | 86.851 | ENSMGAG00000001112 | PRDM10 | 100 | 82.235 | Meleagris_gallopavo |
| ENSG00000170325 | PRDM10 | 100 | 96.810 | ENSMICG00000013888 | PRDM10 | 100 | 96.810 | Microcebus_murinus |
| ENSG00000170325 | PRDM10 | 100 | 92.971 | ENSMOCG00000021853 | Prdm10 | 100 | 90.878 | Microtus_ochrogaster |
| ENSG00000170325 | PRDM10 | 100 | 87.115 | ENSMODG00000004979 | PRDM10 | 99 | 87.443 | Monodelphis_domestica |
| ENSG00000170325 | PRDM10 | 100 | 94.335 | ENSMUSG00000042496 | Prdm10 | 100 | 93.834 | Mus_musculus |
| ENSG00000170325 | PRDM10 | 99 | 94.240 | MGP_SPRETEiJ_G0033061 | - | 100 | 92.599 | Mus_spretus |
| ENSG00000170325 | PRDM10 | 100 | 94.896 | ENSMPUG00000008711 | PRDM10 | 99 | 94.896 | Mustela_putorius_furo |
| ENSG00000170325 | PRDM10 | 100 | 93.362 | ENSMLUG00000013051 | PRDM10 | 100 | 93.362 | Myotis_lucifugus |
| ENSG00000170325 | PRDM10 | 99 | 96.647 | ENSNGAG00000000580 | Prdm10 | 100 | 95.156 | Nannospalax_galili |
| ENSG00000170325 | PRDM10 | 100 | 99.281 | ENSNLEG00000007488 | PRDM10 | 100 | 98.707 | Nomascus_leucogenys |
| ENSG00000170325 | PRDM10 | 100 | 90.508 | ENSODEG00000005309 | PRDM10 | 99 | 94.805 | Octodon_degus |
| ENSG00000170325 | PRDM10 | 100 | 82.200 | ENSOANG00000000577 | - | 100 | 79.745 | Ornithorhynchus_anatinus |
| ENSG00000170325 | PRDM10 | 100 | 93.772 | ENSOCUG00000003736 | PRDM10 | 100 | 93.685 | Oryctolagus_cuniculus |
| ENSG00000170325 | PRDM10 | 100 | 95.000 | ENSOGAG00000010723 | PRDM10 | 100 | 94.655 | Otolemur_garnettii |
| ENSG00000170325 | PRDM10 | 100 | 93.772 | ENSOARG00000014507 | PRDM10 | 100 | 93.166 | Ovis_aries |
| ENSG00000170325 | PRDM10 | 100 | 99.307 | ENSPPAG00000042881 | PRDM10 | 100 | 99.225 | Pan_paniscus |
| ENSG00000170325 | PRDM10 | 100 | 94.569 | ENSPPRG00000007002 | PRDM10 | 100 | 94.569 | Panthera_pardus |
| ENSG00000170325 | PRDM10 | 100 | 94.655 | ENSPTIG00000013331 | PRDM10 | 100 | 94.655 | Panthera_tigris_altaica |
| ENSG00000170325 | PRDM10 | 100 | 100.000 | ENSPTRG00000004477 | PRDM10 | 100 | 99.225 | Pan_troglodytes |
| ENSG00000170325 | PRDM10 | 100 | 99.281 | ENSPANG00000017470 | PRDM10 | 100 | 98.793 | Papio_anubis |
| ENSG00000170325 | PRDM10 | 100 | 92.873 | ENSPEMG00000014985 | Prdm10 | 100 | 91.249 | Peromyscus_maniculatus_bairdii |
| ENSG00000170325 | PRDM10 | 100 | 99.422 | ENSPPYG00000004061 | PRDM10 | 100 | 99.394 | Pongo_abelii |
| ENSG00000170325 | PRDM10 | 93 | 87.035 | ENSPCAG00000014514 | PRDM10 | 99 | 85.888 | Procavia_capensis |
| ENSG00000170325 | PRDM10 | 100 | 82.631 | ENSPCOG00000023406 | PRDM10 | 100 | 82.631 | Propithecus_coquereli |
| ENSG00000170325 | PRDM10 | 100 | 94.914 | ENSPVAG00000012764 | PRDM10 | 100 | 95.000 | Pteropus_vampyrus |
| ENSG00000170325 | PRDM10 | 99 | 94.792 | ENSRNOG00000007853 | Prdm10 | 100 | 92.919 | Rattus_norvegicus |
| ENSG00000170325 | PRDM10 | 100 | 99.281 | ENSRBIG00000030924 | PRDM10 | 100 | 99.052 | Rhinopithecus_bieti |
| ENSG00000170325 | PRDM10 | 100 | 99.075 | ENSRROG00000034334 | PRDM10 | 100 | 99.052 | Rhinopithecus_roxellana |
| ENSG00000170325 | PRDM10 | 100 | 98.497 | ENSSBOG00000008773 | PRDM10 | 100 | 98.190 | Saimiri_boliviensis_boliviensis |
| ENSG00000170325 | PRDM10 | 100 | 87.743 | ENSSHAG00000004503 | PRDM10 | 99 | 87.360 | Sarcophilus_harrisii |
| ENSG00000170325 | PRDM10 | 74 | 100.000 | ENSSARG00000013953 | - | 68 | 100.000 | Sorex_araneus |
| ENSG00000170325 | PRDM10 | 100 | 85.763 | ENSSPUG00000001178 | PRDM10 | 99 | 83.673 | Sphenodon_punctatus |
| ENSG00000170325 | PRDM10 | 100 | 93.064 | ENSSSCG00000015243 | PRDM10 | 100 | 93.287 | Sus_scrofa |
| ENSG00000170325 | PRDM10 | 99 | 85.747 | ENSTGUG00000000530 | PRDM10 | 100 | 83.691 | Taeniopygia_guttata |
| ENSG00000170325 | PRDM10 | 93 | 98.204 | ENSTBEG00000007221 | PRDM10 | 94 | 95.133 | Tupaia_belangeri |
| ENSG00000170325 | PRDM10 | 100 | 92.175 | ENSTTRG00000009189 | PRDM10 | 100 | 90.690 | Tursiops_truncatus |
| ENSG00000170325 | PRDM10 | 91 | 96.256 | ENSUAMG00000018832 | PRDM10 | 95 | 96.256 | Ursus_americanus |
| ENSG00000170325 | PRDM10 | 100 | 94.451 | ENSUMAG00000003167 | PRDM10 | 99 | 94.995 | Ursus_maritimus |
| ENSG00000170325 | PRDM10 | 100 | 94.291 | ENSVVUG00000019878 | PRDM10 | 100 | 94.291 | Vulpes_vulpes |
| ENSG00000170325 | PRDM10 | 99 | 72.269 | ENSXETG00000032173 | PRDM10 | 100 | 69.417 | Xenopus_tropicalis |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding | 21873635. | IBA | Function |
| GO:0000981 | DNA-binding transcription factor activity, RNA polymerase II-specific | - | ISA | Function |
| GO:0005515 | protein binding | 25416956. | IPI | Function |
| GO:0005634 | nucleus | 21873635. | IBA | Component |
| GO:0006357 | regulation of transcription by RNA polymerase II | - | IEA | Process |
| GO:0008168 | methyltransferase activity | - | IEA | Function |
| GO:0010468 | regulation of gene expression | 21873635. | IBA | Process |
| GO:0032259 | methylation | - | IEA | Process |
| GO:0035097 | histone methyltransferase complex | 21873635. | IBA | Component |
| GO:0046872 | metal ion binding | - | IEA | Function |