
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000377796 | Methyltransf_31 | PF13847.6 | 4.4e-11 | 1 | 1 |
| ENSP00000482867 | Methyltransf_31 | PF13847.6 | 5.9e-11 | 1 | 1 |
| ENSP00000377796 | Methyltransf_8 | PF05148.15 | 0.00023 | 1 | 1 |
| ENSP00000482867 | Methyltransf_8 | PF05148.15 | 0.00028 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000614691 | METTL7B-202 | 1368 | - | ENSP00000482867 | 276 (aa) | - | A0A087WZT2 |
| ENST00000394252 | METTL7B-201 | 1506 | - | ENSP00000377796 | 244 (aa) | - | Q6UX53 |
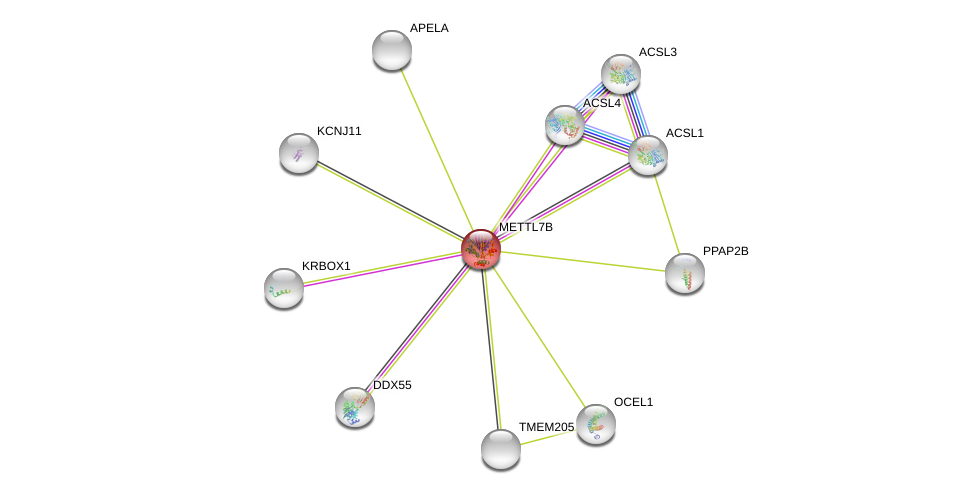
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000170439 | METTL7B | 91 | 84.753 | ENSAMEG00000011933 | METTL7B | 91 | 84.753 | Ailuropoda_melanoleuca |
| ENSG00000170439 | METTL7B | 91 | 91.480 | ENSANAG00000036477 | METTL7B | 91 | 91.480 | Aotus_nancymaae |
| ENSG00000170439 | METTL7B | 92 | 85.778 | ENSBTAG00000012896 | METTL7B | 91 | 85.650 | Bos_taurus |
| ENSG00000170439 | METTL7B | 91 | 89.641 | ENSCJAG00000008201 | METTL7B | 92 | 89.412 | Callithrix_jacchus |
| ENSG00000170439 | METTL7B | 91 | 87.444 | ENSCAFG00000031038 | METTL7B | 91 | 87.444 | Canis_familiaris |
| ENSG00000170439 | METTL7B | 91 | 86.937 | ENSCAFG00020026120 | METTL7B | 91 | 82.917 | Canis_lupus_dingo |
| ENSG00000170439 | METTL7B | 92 | 87.556 | ENSCHIG00000027137 | METTL7B | 91 | 87.444 | Capra_hircus |
| ENSG00000170439 | METTL7B | 91 | 91.071 | ENSTSYG00000007994 | METTL7B | 91 | 91.071 | Carlito_syrichta |
| ENSG00000170439 | METTL7B | 89 | 82.488 | ENSCPOG00000014972 | METTL7B | 91 | 82.063 | Cavia_porcellus |
| ENSG00000170439 | METTL7B | 91 | 94.170 | ENSCCAG00000028514 | METTL7B | 91 | 94.170 | Cebus_capucinus |
| ENSG00000170439 | METTL7B | 91 | 95.516 | ENSCATG00000041077 | METTL7B | 91 | 95.516 | Cercocebus_atys |
| ENSG00000170439 | METTL7B | 89 | 87.558 | ENSCLAG00000016551 | METTL7B | 91 | 86.996 | Chinchilla_lanigera |
| ENSG00000170439 | METTL7B | 91 | 95.964 | ENSCSAG00000005338 | METTL7B | 91 | 95.964 | Chlorocebus_sabaeus |
| ENSG00000170439 | METTL7B | 91 | 95.067 | ENSCANG00000039775 | METTL7B | 91 | 95.067 | Colobus_angolensis_palliatus |
| ENSG00000170439 | METTL7B | 90 | 84.091 | ENSCGRG00001020267 | Mettl7b | 91 | 83.857 | Cricetulus_griseus_chok1gshd |
| ENSG00000170439 | METTL7B | 90 | 84.091 | ENSCGRG00000015365 | Mettl7b | 91 | 83.857 | Cricetulus_griseus_crigri |
| ENSG00000170439 | METTL7B | 99 | 50.617 | ENSCSEG00000003205 | - | 87 | 52.113 | Cynoglossus_semilaevis |
| ENSG00000170439 | METTL7B | 91 | 89.686 | ENSDNOG00000046100 | METTL7B | 100 | 89.344 | Dasypus_novemcinctus |
| ENSG00000170439 | METTL7B | 100 | 81.148 | ENSDORG00000014149 | Mettl7b | 100 | 81.148 | Dipodomys_ordii |
| ENSG00000170439 | METTL7B | 80 | 81.959 | ENSETEG00000009099 | METTL7B | 100 | 81.959 | Echinops_telfairi |
| ENSG00000170439 | METTL7B | 91 | 85.973 | ENSEASG00005001861 | METTL7B | 90 | 83.475 | Equus_asinus_asinus |
| ENSG00000170439 | METTL7B | 91 | 86.099 | ENSECAG00000011720 | METTL7B | 91 | 86.099 | Equus_caballus |
| ENSG00000170439 | METTL7B | 91 | 88.341 | ENSFCAG00000039385 | METTL7B | 91 | 88.341 | Felis_catus |
| ENSG00000170439 | METTL7B | 89 | 84.332 | ENSFDAG00000014366 | METTL7B | 91 | 83.857 | Fukomys_damarensis |
| ENSG00000170439 | METTL7B | 100 | 51.639 | ENSGMOG00000002697 | - | 88 | 53.738 | Gadus_morhua |
| ENSG00000170439 | METTL7B | 100 | 100.000 | ENSGGOG00000008581 | METTL7B | 100 | 100.000 | Gorilla_gorilla |
| ENSG00000170439 | METTL7B | 89 | 81.567 | ENSHGLG00000006754 | METTL7B | 91 | 81.166 | Heterocephalus_glaber_female |
| ENSG00000170439 | METTL7B | 89 | 81.567 | ENSHGLG00100016921 | METTL7B | 91 | 81.166 | Heterocephalus_glaber_male |
| ENSG00000170439 | METTL7B | 90 | 87.215 | ENSJJAG00000015629 | Mettl7b | 100 | 86.885 | Jaculus_jaculus |
| ENSG00000170439 | METTL7B | 91 | 86.937 | ENSLAFG00000032588 | METTL7B | 91 | 86.547 | Loxodonta_africana |
| ENSG00000170439 | METTL7B | 91 | 95.516 | ENSMFAG00000041138 | METTL7B | 91 | 95.516 | Macaca_fascicularis |
| ENSG00000170439 | METTL7B | 91 | 95.516 | ENSMMUG00000013225 | METTL7B | 91 | 95.516 | Macaca_mulatta |
| ENSG00000170439 | METTL7B | 91 | 95.516 | ENSMNEG00000030037 | METTL7B | 91 | 95.516 | Macaca_nemestrina |
| ENSG00000170439 | METTL7B | 91 | 94.619 | ENSMLEG00000033453 | METTL7B | 91 | 94.619 | Mandrillus_leucophaeus |
| ENSG00000170439 | METTL7B | 100 | 84.426 | ENSMAUG00000015292 | Mettl7b | 91 | 84.753 | Mesocricetus_auratus |
| ENSG00000170439 | METTL7B | 100 | 80.738 | ENSMOCG00000008376 | Mettl7b | 91 | 80.269 | Microtus_ochrogaster |
| ENSG00000170439 | METTL7B | 100 | 77.459 | ENSMODG00000001722 | METTL7B | 91 | 78.475 | Monodelphis_domestica |
| ENSG00000170439 | METTL7B | 90 | 83.562 | MGP_CAROLIEiJ_G0015964 | Mettl7b | 91 | 83.408 | Mus_caroli |
| ENSG00000170439 | METTL7B | 90 | 83.562 | ENSMUSG00000025347 | Mettl7b | 91 | 83.408 | Mus_musculus |
| ENSG00000170439 | METTL7B | 90 | 82.648 | MGP_PahariEiJ_G0031120 | Mettl7b | 91 | 82.511 | Mus_pahari |
| ENSG00000170439 | METTL7B | 90 | 84.018 | MGP_SPRETEiJ_G0016789 | Mettl7b | 91 | 83.857 | Mus_spretus |
| ENSG00000170439 | METTL7B | 90 | 85.845 | ENSMPUG00000000852 | METTL7B | 91 | 85.650 | Mustela_putorius_furo |
| ENSG00000170439 | METTL7B | 90 | 87.671 | ENSNGAG00000018609 | Mettl7b | 91 | 87.444 | Nannospalax_galili |
| ENSG00000170439 | METTL7B | 100 | 98.770 | ENSNLEG00000017612 | METTL7B | 100 | 98.770 | Nomascus_leucogenys |
| ENSG00000170439 | METTL7B | 100 | 79.098 | ENSMEUG00000000400 | METTL7B | 91 | 80.717 | Notamacropus_eugenii |
| ENSG00000170439 | METTL7B | 89 | 87.097 | ENSODEG00000004695 | METTL7B | 91 | 86.547 | Octodon_degus |
| ENSG00000170439 | METTL7B | 91 | 86.996 | ENSOCUG00000023347 | METTL7B | 91 | 86.996 | Oryctolagus_cuniculus |
| ENSG00000170439 | METTL7B | 92 | 92.444 | ENSOGAG00000010513 | METTL7B | 91 | 92.377 | Otolemur_garnettii |
| ENSG00000170439 | METTL7B | 90 | 88.128 | ENSOARG00000011505 | METTL7B | 93 | 87.892 | Ovis_aries |
| ENSG00000170439 | METTL7B | 100 | 100.000 | ENSPPAG00000029723 | METTL7B | 100 | 100.000 | Pan_paniscus |
| ENSG00000170439 | METTL7B | 91 | 88.789 | ENSPPRG00000023193 | METTL7B | 91 | 88.789 | Panthera_pardus |
| ENSG00000170439 | METTL7B | 91 | 88.341 | ENSPTIG00000012796 | METTL7B | 91 | 88.341 | Panthera_tigris_altaica |
| ENSG00000170439 | METTL7B | 100 | 100.000 | ENSPTRG00000005058 | METTL7B | 100 | 100.000 | Pan_troglodytes |
| ENSG00000170439 | METTL7B | 91 | 95.516 | ENSPANG00000015541 | METTL7B | 91 | 95.516 | Papio_anubis |
| ENSG00000170439 | METTL7B | 90 | 83.562 | ENSPEMG00000001761 | Mettl7b | 91 | 83.408 | Peromyscus_maniculatus_bairdii |
| ENSG00000170439 | METTL7B | 100 | 79.098 | ENSPCIG00000024669 | METTL7B | 91 | 80.717 | Phascolarctos_cinereus |
| ENSG00000170439 | METTL7B | 100 | 97.131 | ENSPPYG00000004625 | METTL7B | 100 | 97.131 | Pongo_abelii |
| ENSG00000170439 | METTL7B | 91 | 84.234 | ENSPCAG00000008996 | METTL7B | 91 | 83.857 | Procavia_capensis |
| ENSG00000170439 | METTL7B | 84 | 76.699 | ENSPCOG00000021279 | METTL7B | 91 | 76.699 | Propithecus_coquereli |
| ENSG00000170439 | METTL7B | 90 | 80.543 | ENSPVAG00000017745 | METTL7B | 91 | 80.889 | Pteropus_vampyrus |
| ENSG00000170439 | METTL7B | 90 | 82.648 | ENSRNOG00000007927 | Mettl7b | 91 | 82.511 | Rattus_norvegicus |
| ENSG00000170439 | METTL7B | 91 | 94.170 | ENSRBIG00000028549 | METTL7B | 91 | 94.170 | Rhinopithecus_bieti |
| ENSG00000170439 | METTL7B | 91 | 94.170 | ENSRROG00000043640 | METTL7B | 91 | 94.170 | Rhinopithecus_roxellana |
| ENSG00000170439 | METTL7B | 91 | 93.722 | ENSSBOG00000013267 | METTL7B | 91 | 93.722 | Saimiri_boliviensis_boliviensis |
| ENSG00000170439 | METTL7B | 91 | 83.857 | ENSSSCG00000036547 | METTL7B | 91 | 83.857 | Sus_scrofa |
| ENSG00000170439 | METTL7B | 80 | 92.268 | ENSTBEG00000015579 | METTL7B | 100 | 92.268 | Tupaia_belangeri |
| ENSG00000170439 | METTL7B | 92 | 88.444 | ENSTTRG00000009630 | METTL7B | 91 | 88.341 | Tursiops_truncatus |
| ENSG00000170439 | METTL7B | 91 | 84.753 | ENSUAMG00000015092 | METTL7B | 91 | 84.753 | Ursus_americanus |
| ENSG00000170439 | METTL7B | 91 | 84.753 | ENSUMAG00000015411 | METTL7B | 91 | 84.753 | Ursus_maritimus |
| ENSG00000170439 | METTL7B | 92 | 89.778 | ENSVPAG00000009509 | METTL7B | 91 | 90.135 | Vicugna_pacos |
| ENSG00000170439 | METTL7B | 91 | 86.996 | ENSVVUG00000023831 | METTL7B | 91 | 86.996 | Vulpes_vulpes |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0008168 | methyltransferase activity | - | IEA | Function |
| GO:0032259 | methylation | - | IEA | Process |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| ACC | |||||||
| ACC | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| PAAD | |||||||
| READ | |||||||
| READ | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| DLBC | |||
| PCPG | |||
| READ |
| Cancer | P-value | Q-value |
|---|---|---|
| THYM | ||
| STAD | ||
| SARC | ||
| HNSC | ||
| SKCM | ||
| LUSC | ||
| PAAD | ||
| LAML | ||
| UCEC | ||
| GBM | ||
| THCA | ||
| UVM |