
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000306080 | SpoU_methylase | PF00588.19 | 8.7e-21 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000574509 | MRM3-203 | 727 | - | ENSP00000458328 | 110 (aa) | - | I3L0T6 |
| ENST00000574916 | MRM3-204 | 770 | - | - | - (aa) | - | - |
| ENST00000571157 | MRM3-202 | 572 | - | ENSP00000460957 | 177 (aa) | - | I3L443 |
| ENST00000304478 | MRM3-201 | 1809 | - | ENSP00000306080 | 420 (aa) | - | Q9HC36 |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000171861 | Diabetes Mellitus, Type 2 | 4E-6 | 23300278 |
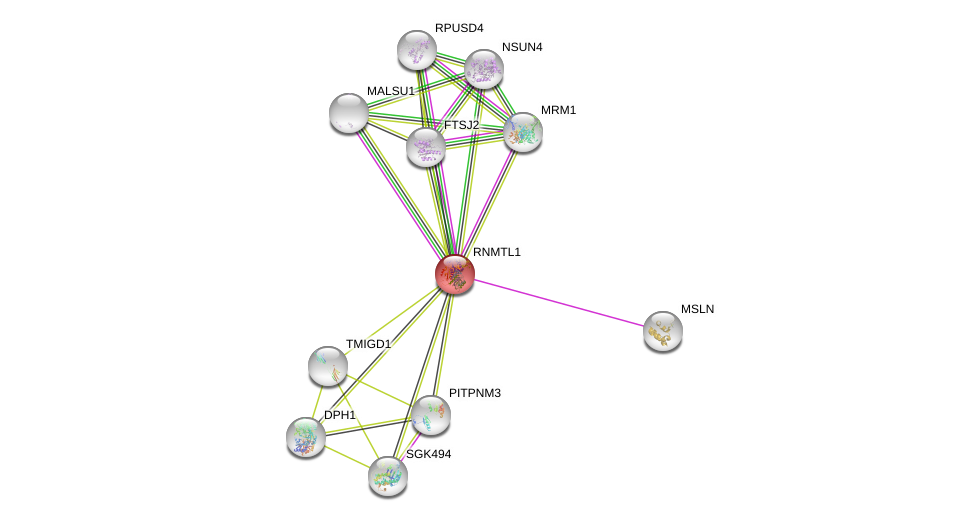
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000171861 | MRM3 | 95 | 90.058 | ENSAMEG00000008346 | MRM3 | 100 | 83.333 | Ailuropoda_melanoleuca |
| ENSG00000171861 | MRM3 | 95 | 94.643 | ENSANAG00000014643 | MRM3 | 97 | 90.120 | Aotus_nancymaae |
| ENSG00000171861 | MRM3 | 95 | 95.266 | ENSCJAG00000021701 | MRM3 | 97 | 85.301 | Callithrix_jacchus |
| ENSG00000171861 | MRM3 | 95 | 91.124 | ENSCAFG00000019117 | MRM3 | 100 | 83.924 | Canis_familiaris |
| ENSG00000171861 | MRM3 | 95 | 91.124 | ENSCAFG00020010972 | MRM3 | 100 | 83.924 | Canis_lupus_dingo |
| ENSG00000171861 | MRM3 | 93 | 91.463 | ENSCHIG00000025241 | MRM3 | 99 | 84.048 | Capra_hircus |
| ENSG00000171861 | MRM3 | 95 | 91.124 | ENSTSYG00000006075 | MRM3 | 100 | 85.986 | Carlito_syrichta |
| ENSG00000171861 | MRM3 | 66 | 88.172 | ENSCAPG00000011399 | MRM3 | 99 | 88.172 | Cavia_aperea |
| ENSG00000171861 | MRM3 | 95 | 88.757 | ENSCPOG00000026540 | MRM3 | 99 | 84.198 | Cavia_porcellus |
| ENSG00000171861 | MRM3 | 95 | 94.675 | ENSCCAG00000038105 | MRM3 | 99 | 89.639 | Cebus_capucinus |
| ENSG00000171861 | MRM3 | 100 | 95.000 | ENSCATG00000032099 | MRM3 | 100 | 95.000 | Cercocebus_atys |
| ENSG00000171861 | MRM3 | 100 | 83.294 | ENSCLAG00000008589 | MRM3 | 100 | 84.000 | Chinchilla_lanigera |
| ENSG00000171861 | MRM3 | 100 | 95.000 | ENSCSAG00000014247 | MRM3 | 100 | 95.000 | Chlorocebus_sabaeus |
| ENSG00000171861 | MRM3 | 95 | 88.166 | ENSCHOG00000011403 | MRM3 | 100 | 82.619 | Choloepus_hoffmanni |
| ENSG00000171861 | MRM3 | 100 | 93.571 | ENSCANG00000040883 | MRM3 | 100 | 93.571 | Colobus_angolensis_palliatus |
| ENSG00000171861 | MRM3 | 95 | 91.667 | ENSCGRG00001014632 | Mrm3 | 100 | 83.333 | Cricetulus_griseus_chok1gshd |
| ENSG00000171861 | MRM3 | 95 | 91.667 | ENSCGRG00000016619 | Mrm3 | 100 | 83.333 | Cricetulus_griseus_crigri |
| ENSG00000171861 | MRM3 | 95 | 87.574 | ENSDNOG00000004294 | MRM3 | 99 | 83.055 | Dasypus_novemcinctus |
| ENSG00000171861 | MRM3 | 93 | 86.667 | ENSDORG00000007555 | Mrm3 | 99 | 80.976 | Dipodomys_ordii |
| ENSG00000171861 | MRM3 | 95 | 90.533 | ENSEASG00005009647 | MRM3 | 100 | 85.238 | Equus_asinus_asinus |
| ENSG00000171861 | MRM3 | 95 | 91.124 | ENSECAG00000024378 | MRM3 | 92 | 86.241 | Equus_caballus |
| ENSG00000171861 | MRM3 | 95 | 90.476 | ENSFCAG00000001673 | MRM3 | 100 | 83.610 | Felis_catus |
| ENSG00000171861 | MRM3 | 100 | 100.000 | ENSGGOG00000015518 | MRM3 | 100 | 100.000 | Gorilla_gorilla |
| ENSG00000171861 | MRM3 | 99 | 85.071 | ENSHGLG00000009544 | MRM3 | 100 | 85.071 | Heterocephalus_glaber_female |
| ENSG00000171861 | MRM3 | 99 | 85.442 | ENSHGLG00100019310 | MRM3 | 100 | 85.442 | Heterocephalus_glaber_male |
| ENSG00000171861 | MRM3 | 100 | 85.238 | ENSSTOG00000016426 | MRM3 | 100 | 85.238 | Ictidomys_tridecemlineatus |
| ENSG00000171861 | MRM3 | 95 | 86.391 | ENSJJAG00000024189 | Mrm3 | 100 | 78.095 | Jaculus_jaculus |
| ENSG00000171861 | MRM3 | 86 | 68.627 | ENSLACG00000007157 | MRM3 | 95 | 54.462 | Latimeria_chalumnae |
| ENSG00000171861 | MRM3 | 95 | 87.719 | ENSLAFG00000016652 | MRM3 | 100 | 82.547 | Loxodonta_africana |
| ENSG00000171861 | MRM3 | 100 | 94.048 | ENSMFAG00000045382 | MRM3 | 100 | 94.048 | Macaca_fascicularis |
| ENSG00000171861 | MRM3 | 100 | 93.810 | ENSMNEG00000032873 | MRM3 | 100 | 93.810 | Macaca_nemestrina |
| ENSG00000171861 | MRM3 | 100 | 94.524 | ENSMLEG00000034316 | MRM3 | 100 | 94.524 | Mandrillus_leucophaeus |
| ENSG00000171861 | MRM3 | 95 | 89.941 | ENSMAUG00000018823 | Mrm3 | 100 | 82.619 | Mesocricetus_auratus |
| ENSG00000171861 | MRM3 | 100 | 86.429 | ENSMICG00000031420 | MRM3 | 100 | 86.429 | Microcebus_murinus |
| ENSG00000171861 | MRM3 | 95 | 91.124 | ENSMOCG00000018749 | Mrm3 | 100 | 82.619 | Microtus_ochrogaster |
| ENSG00000171861 | MRM3 | 91 | 85.093 | ENSMODG00000010029 | MRM3 | 93 | 72.115 | Monodelphis_domestica |
| ENSG00000171861 | MRM3 | 95 | 90.533 | MGP_CAROLIEiJ_G0016758 | Mrm3 | 97 | 84.236 | Mus_caroli |
| ENSG00000171861 | MRM3 | 95 | 89.941 | ENSMUSG00000038046 | Mrm3 | 97 | 83.251 | Mus_musculus |
| ENSG00000171861 | MRM3 | 95 | 90.533 | MGP_PahariEiJ_G0017895 | Mrm3 | 100 | 82.143 | Mus_pahari |
| ENSG00000171861 | MRM3 | 95 | 89.941 | MGP_SPRETEiJ_G0017601 | Mrm3 | 97 | 83.251 | Mus_spretus |
| ENSG00000171861 | MRM3 | 95 | 91.716 | ENSMPUG00000013793 | MRM3 | 100 | 80.601 | Mustela_putorius_furo |
| ENSG00000171861 | MRM3 | 99 | 83.333 | ENSMLUG00000001504 | MRM3 | 100 | 83.333 | Myotis_lucifugus |
| ENSG00000171861 | MRM3 | 95 | 92.899 | ENSNGAG00000011304 | Mrm3 | 100 | 85.714 | Nannospalax_galili |
| ENSG00000171861 | MRM3 | 100 | 95.952 | ENSNLEG00000033176 | MRM3 | 100 | 95.952 | Nomascus_leucogenys |
| ENSG00000171861 | MRM3 | 77 | 86.131 | ENSMEUG00000012224 | MRM3 | 99 | 82.000 | Notamacropus_eugenii |
| ENSG00000171861 | MRM3 | 100 | 75.238 | ENSODEG00000009411 | MRM3 | 100 | 75.238 | Octodon_degus |
| ENSG00000171861 | MRM3 | 77 | 77.372 | ENSOANG00000009926 | MRM3 | 97 | 73.684 | Ornithorhynchus_anatinus |
| ENSG00000171861 | MRM3 | 95 | 85.207 | ENSOCUG00000002219 | MRM3 | 100 | 78.673 | Oryctolagus_cuniculus |
| ENSG00000171861 | MRM3 | 99 | 86.190 | ENSOGAG00000001360 | MRM3 | 100 | 86.190 | Otolemur_garnettii |
| ENSG00000171861 | MRM3 | 95 | 88.235 | ENSOARG00000011376 | MRM3 | 99 | 82.938 | Ovis_aries |
| ENSG00000171861 | MRM3 | 100 | 99.524 | ENSPPAG00000038493 | MRM3 | 100 | 99.524 | Pan_paniscus |
| ENSG00000171861 | MRM3 | 95 | 89.881 | ENSPPRG00000008082 | MRM3 | 100 | 82.898 | Panthera_pardus |
| ENSG00000171861 | MRM3 | 95 | 90.476 | ENSPTIG00000021325 | MRM3 | 100 | 83.135 | Panthera_tigris_altaica |
| ENSG00000171861 | MRM3 | 100 | 99.762 | ENSPTRG00000008512 | MRM3 | 100 | 99.762 | Pan_troglodytes |
| ENSG00000171861 | MRM3 | 100 | 95.238 | ENSPANG00000024613 | MRM3 | 100 | 95.238 | Papio_anubis |
| ENSG00000171861 | MRM3 | 86 | 62.745 | ENSPKIG00000012704 | MRM3 | 97 | 47.921 | Paramormyrops_kingsleyae |
| ENSG00000171861 | MRM3 | 95 | 88.824 | ENSPEMG00000022505 | Mrm3 | 100 | 83.135 | Peromyscus_maniculatus_bairdii |
| ENSG00000171861 | MRM3 | 86 | 55.195 | ENSPMAG00000001598 | MRM3 | 99 | 45.858 | Petromyzon_marinus |
| ENSG00000171861 | MRM3 | 95 | 82.738 | ENSPCIG00000025698 | MRM3 | 88 | 73.684 | Phascolarctos_cinereus |
| ENSG00000171861 | MRM3 | 95 | 88.166 | ENSPCAG00000008042 | MRM3 | 100 | 81.473 | Procavia_capensis |
| ENSG00000171861 | MRM3 | 100 | 86.936 | ENSPCOG00000012539 | MRM3 | 100 | 86.936 | Propithecus_coquereli |
| ENSG00000171861 | MRM3 | 95 | 88.757 | ENSPVAG00000011709 | MRM3 | 100 | 83.135 | Pteropus_vampyrus |
| ENSG00000171861 | MRM3 | 86 | 66.013 | ENSPNAG00000019511 | MRM3 | 94 | 46.729 | Pygocentrus_nattereri |
| ENSG00000171861 | MRM3 | 95 | 92.899 | ENSRNOG00000037238 | Mrm3 | 100 | 82.619 | Rattus_norvegicus |
| ENSG00000171861 | MRM3 | 99 | 93.317 | ENSRBIG00000044238 | MRM3 | 99 | 93.317 | Rhinopithecus_bieti |
| ENSG00000171861 | MRM3 | 99 | 94.033 | ENSRROG00000041720 | MRM3 | 98 | 94.033 | Rhinopithecus_roxellana |
| ENSG00000171861 | MRM3 | 95 | 94.083 | ENSSBOG00000029574 | MRM3 | 99 | 89.157 | Saimiri_boliviensis_boliviensis |
| ENSG00000171861 | MRM3 | 67 | 82.203 | ENSSHAG00000011624 | MRM3 | 90 | 77.739 | Sarcophilus_harrisii |
| ENSG00000171861 | MRM3 | 91 | 85.714 | ENSSARG00000006188 | MRM3 | 100 | 77.889 | Sorex_araneus |
| ENSG00000171861 | MRM3 | 95 | 89.941 | ENSSSCG00000017806 | MRM3 | 100 | 82.506 | Sus_scrofa |
| ENSG00000171861 | MRM3 | 100 | 82.033 | ENSTTRG00000003844 | MRM3 | 100 | 82.270 | Tursiops_truncatus |
| ENSG00000171861 | MRM3 | 95 | 91.716 | ENSUAMG00000013654 | MRM3 | 100 | 83.019 | Ursus_americanus |
| ENSG00000171861 | MRM3 | 95 | 91.716 | ENSUMAG00000010052 | MRM3 | 100 | 83.255 | Ursus_maritimus |
| ENSG00000171861 | MRM3 | 86 | 67.105 | ENSXETG00000022451 | mrm3 | 98 | 54.940 | Xenopus_tropicalis |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000451 | rRNA 2'-O-methylation | - | TAS | Process |
| GO:0003723 | RNA binding | 22658674.22681889. | HDA | Function |
| GO:0005515 | protein binding | 25416956. | IPI | Function |
| GO:0005739 | mitochondrion | - | IDA | Component |
| GO:0005759 | mitochondrial matrix | - | TAS | Component |
| GO:0070039 | rRNA (guanosine-2'-O-)-methyltransferase activity | 24036117.25074936. | EXP | Function |