
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000590533 | THOP1-210 | 616 | - | - | - (aa) | - | - |
| ENST00000586677 | THOP1-205 | 1872 | XM_011528228 | ENSP00000467226 | 568 (aa) | XP_011526530 | K7EP46 |
| ENST00000586780 | THOP1-206 | 421 | - | - | - (aa) | - | - |
| ENST00000591149 | THOP1-212 | 789 | - | - | - (aa) | - | - |
| ENST00000591363 | THOP1-213 | 639 | - | ENSP00000466653 | 204 (aa) | - | K7EMU4 |
| ENST00000592639 | THOP1-214 | 431 | - | - | - (aa) | - | - |
| ENST00000395212 | THOP1-202 | 1877 | - | ENSP00000378638 | 258 (aa) | - | P52888 |
| ENST00000587401 | THOP1-207 | 702 | - | ENSP00000465895 | 216 (aa) | - | K7EL32 |
| ENST00000585673 | THOP1-204 | 784 | - | - | - (aa) | - | - |
| ENST00000589087 | THOP1-209 | 2977 | - | - | - (aa) | - | - |
| ENST00000587468 | THOP1-208 | 671 | - | ENSP00000464786 | 169 (aa) | - | K7EIK4 |
| ENST00000590970 | THOP1-211 | 1104 | - | ENSP00000465855 | 260 (aa) | - | K7EL02 |
| ENST00000585338 | THOP1-203 | 761 | - | ENSP00000465545 | 176 (aa) | - | K7EKB6 |
| ENST00000307741 | THOP1-201 | 4804 | - | ENSP00000304467 | 689 (aa) | - | P52888 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000172009 | rs941408 | 19 | 2814183 | T | Total cholesterol levels | 28334899 | [0.012-0.028] unit increase (EA Beta values) | 0.02 | EFO_0004574 |
| ENSG00000172009 | rs941408 | 19 | 2814183 | C | Low density lipoprotein cholesterol levels | 29507422 | unit decrease | 0.018 | EFO_0004611 |
| ENSG00000172009 | rs941408 | 19 | 2814183 | C | Total cholesterol levels | 29507422 | unit decrease | 0.021 | EFO_0004574 |
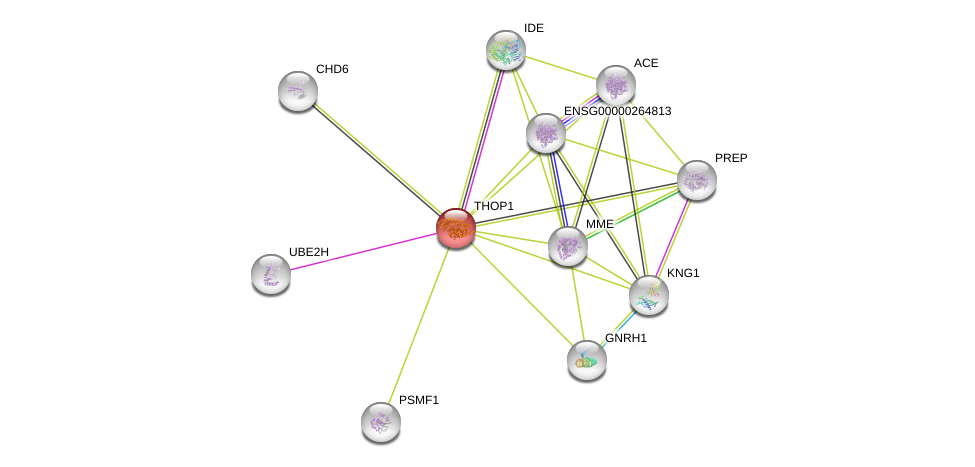
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000172009 | THOP1 | 99 | 69.048 | ENSG00000123213 | NLN | 99 | 65.050 | Homo_sapiens |
| ENSG00000172009 | THOP1 | 100 | 91.716 | ENSMUSG00000004929 | Thop1 | 99 | 89.605 | Mus_musculus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000209 | protein polyubiquitination | - | TAS | Process |
| GO:0004222 | metalloendopeptidase activity | 21873635. | IBA | Function |
| GO:0004222 | metalloendopeptidase activity | - | TAS | Function |
| GO:0005758 | mitochondrial intermembrane space | 21873635. | IBA | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0006508 | proteolysis | 21873635. | IBA | Process |
| GO:0006518 | peptide metabolic process | 21873635. | IBA | Process |
| GO:0042277 | peptide binding | - | IEA | Function |
| GO:0046872 | metal ion binding | - | IEA | Function |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| ACC | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| KIRP | |||
| LIHC | |||
| PAAD | |||
| READ | |||
| STAD | |||
| THCA | |||
| UCEC | |||
| UCS |
| Cancer | P-value | Q-value |
|---|---|---|
| THYM | ||
| KIRC | ||
| STAD | ||
| MESO | ||
| ACC | ||
| HNSC | ||
| PAAD | ||
| CESC | ||
| READ | ||
| LAML | ||
| UCEC | ||
| DLBC | ||
| LGG | ||
| LUAD | ||
| UVM |