
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000309166 | zf-CCHC | PF00098.23 | 6.7e-07 | 1 | 1 |
| ENSP00000386561 | zf-CCHC | PF00098.23 | 6.7e-07 | 1 | 1 |
| ENSP00000386894 | zf-CCHC | PF00098.23 | 6.7e-07 | 1 | 1 |
| ENSP00000422301 | RRM_5 | PF13893.6 | 3.6e-08 | 1 | 2 |
| ENSP00000422301 | RRM_5 | PF13893.6 | 3.6e-08 | 2 | 2 |
| ENSP00000309166 | RRM_5 | PF13893.6 | 1.4e-07 | 1 | 2 |
| ENSP00000309166 | RRM_5 | PF13893.6 | 1.4e-07 | 2 | 2 |
| ENSP00000386561 | RRM_5 | PF13893.6 | 1.4e-07 | 1 | 2 |
| ENSP00000386561 | RRM_5 | PF13893.6 | 1.4e-07 | 2 | 2 |
| ENSP00000386894 | RRM_5 | PF13893.6 | 1.4e-07 | 1 | 2 |
| ENSP00000386894 | RRM_5 | PF13893.6 | 1.4e-07 | 2 | 2 |
| ENSP00000413497 | RRM_5 | PF13893.6 | 3.5e-06 | 1 | 1 |
| ENSP00000423572 | RRM_5 | PF13893.6 | 3.8e-06 | 1 | 1 |
| ENSP00000381680 | RRM_5 | PF13893.6 | 6.1e-06 | 1 | 1 |
| ENSP00000432150 | RRM_5 | PF13893.6 | 6.1e-06 | 1 | 1 |
| ENSP00000432020 | RRM_5 | PF13893.6 | 6.4e-06 | 1 | 1 |
| ENSP00000435821 | RRM_5 | PF13893.6 | 6.4e-06 | 1 | 1 |
| ENSP00000464349 | RRM_5 | PF13893.6 | 1.4e-05 | 1 | 1 |
| ENSP00000422301 | RRM_1 | PF00076.22 | 2.2e-34 | 1 | 2 |
| ENSP00000422301 | RRM_1 | PF00076.22 | 2.2e-34 | 2 | 2 |
| ENSP00000413497 | RRM_1 | PF00076.22 | 3.4e-34 | 1 | 2 |
| ENSP00000413497 | RRM_1 | PF00076.22 | 3.4e-34 | 2 | 2 |
| ENSP00000381680 | RRM_1 | PF00076.22 | 5.3e-34 | 1 | 2 |
| ENSP00000381680 | RRM_1 | PF00076.22 | 5.3e-34 | 2 | 2 |
| ENSP00000432150 | RRM_1 | PF00076.22 | 5.3e-34 | 1 | 2 |
| ENSP00000432150 | RRM_1 | PF00076.22 | 5.3e-34 | 2 | 2 |
| ENSP00000432020 | RRM_1 | PF00076.22 | 6e-34 | 1 | 2 |
| ENSP00000432020 | RRM_1 | PF00076.22 | 6e-34 | 2 | 2 |
| ENSP00000435821 | RRM_1 | PF00076.22 | 6e-34 | 1 | 2 |
| ENSP00000435821 | RRM_1 | PF00076.22 | 6e-34 | 2 | 2 |
| ENSP00000309166 | RRM_1 | PF00076.22 | 6.9e-34 | 1 | 2 |
| ENSP00000309166 | RRM_1 | PF00076.22 | 6.9e-34 | 2 | 2 |
| ENSP00000386561 | RRM_1 | PF00076.22 | 6.9e-34 | 1 | 2 |
| ENSP00000386561 | RRM_1 | PF00076.22 | 6.9e-34 | 2 | 2 |
| ENSP00000386894 | RRM_1 | PF00076.22 | 6.9e-34 | 1 | 2 |
| ENSP00000386894 | RRM_1 | PF00076.22 | 6.9e-34 | 2 | 2 |
| ENSP00000423572 | RRM_1 | PF00076.22 | 9.5e-34 | 1 | 2 |
| ENSP00000423572 | RRM_1 | PF00076.22 | 9.5e-34 | 2 | 2 |
| ENSP00000464349 | RRM_1 | PF00076.22 | 2.2e-33 | 1 | 2 |
| ENSP00000464349 | RRM_1 | PF00076.22 | 2.2e-33 | 2 | 2 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000398692 | RBM4-203 | 921 | - | ENSP00000381680 | 143 (aa) | - | Q9BWF3 |
| ENST00000310092 | RBM4-201 | 1694 | - | ENSP00000309166 | 364 (aa) | - | Q9BWF3 |
| ENST00000578778 | RBM4-213 | 582 | - | ENSP00000464349 | 155 (aa) | - | J3QRR5 |
| ENST00000510173 | RBM4-208 | 782 | - | ENSP00000422301 | 240 (aa) | - | E9PB51 |
| ENST00000396053 | RBM4-202 | 3029 | - | ENSP00000413497 | 173 (aa) | - | Q9BWF3 |
| ENST00000532968 | RBM4-212 | 757 | - | ENSP00000432020 | 177 (aa) | - | Q9BWF3 |
| ENST00000506523 | RBM4-207 | 654 | - | ENSP00000423572 | 150 (aa) | - | D6R9K7 |
| ENST00000409406 | RBM4-205 | 2321 | - | ENSP00000386894 | 364 (aa) | - | Q9BWF3 |
| ENST00000408993 | RBM4-204 | 1763 | - | ENSP00000386561 | 364 (aa) | - | Q9BWF3 |
| ENST00000530235 | RBM4-211 | 1011 | - | ENSP00000432150 | 143 (aa) | - | Q9BWF3 |
| ENST00000528039 | RBM4-210 | 764 | - | - | - (aa) | - | - |
| ENST00000515838 | RBM4-209 | 943 | - | - | - (aa) | - | - |
| ENST00000483858 | RBM4-206 | 1548 | - | ENSP00000435821 | 177 (aa) | - | Q9BWF3 |
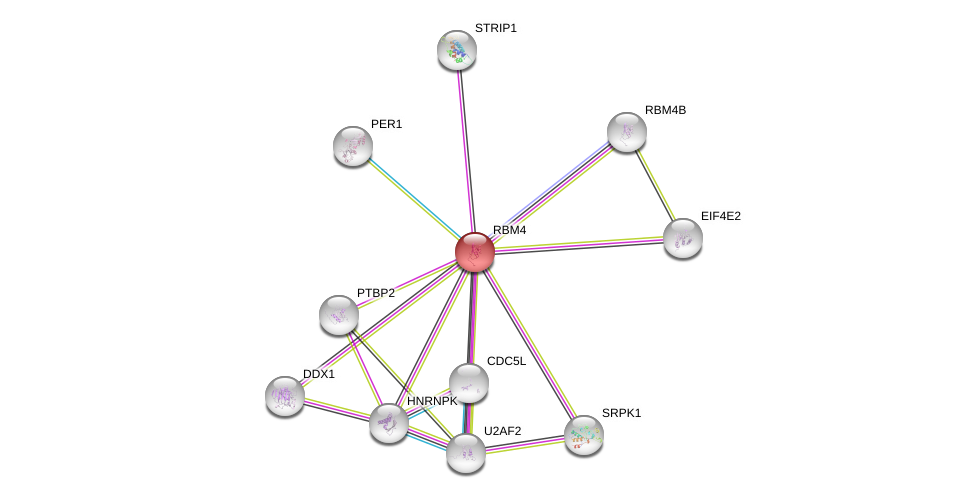
| Ensembl ID | Gene Symbol | Coverage | Identiy | Paralog | Gene Symbol | Coverage | Identiy |
|---|---|---|---|---|---|---|---|
| ENSG00000173933 | RBM4 | 98 | 30.625 | ENSG00000116001 | TIA1 | 85 | 31.081 |
| ENSG00000173933 | RBM4 | 97 | 30.128 | ENSG00000101489 | CELF4 | 52 | 30.065 |
| ENSG00000173933 | RBM4 | 88 | 38.983 | ENSG00000115875 | SRSF7 | 83 | 35.780 |
| ENSG00000173933 | RBM4 | 97 | 98.551 | ENSG00000173914 | RBM4B | 98 | 97.857 |
| ENSG00000173933 | RBM4 | 77 | 35.714 | ENSG00000147140 | NONO | 60 | 32.857 |
| ENSG00000173933 | RBM4 | 50 | 33.750 | ENSG00000213516 | RBMXL1 | 91 | 33.750 |
| ENSG00000173933 | RBM4 | 90 | 30.464 | ENSG00000135486 | HNRNPA1 | 65 | 30.464 |
| ENSG00000173933 | RBM4 | 94 | 31.034 | ENSG00000151923 | TIAL1 | 84 | 30.973 |
| ENSG00000173933 | RBM4 | 59 | 32.143 | ENSG00000124193 | SRSF6 | 62 | 32.143 |
| ENSG00000173933 | RBM4 | 97 | 48.529 | ENSG00000239306 | RBM14 | 94 | 48.529 |
| ENSG00000173933 | RBM4 | 77 | 30.952 | ENSG00000149187 | CELF1 | 79 | 33.333 |
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000173933 | RBM4 | 100 | 100.000 | ENSAMEG00000010914 | RBM4 | 100 | 99.178 | Ailuropoda_melanoleuca |
| ENSG00000173933 | RBM4 | 100 | 98.780 | ENSAMEG00000010906 | RBM4B | 96 | 98.780 | Ailuropoda_melanoleuca |
| ENSG00000173933 | RBM4 | 100 | 96.129 | ENSANAG00000023478 | RBM4B | 100 | 96.129 | Aotus_nancymaae |
| ENSG00000173933 | RBM4 | 100 | 96.503 | ENSANAG00000033986 | - | 96 | 88.679 | Aotus_nancymaae |
| ENSG00000173933 | RBM4 | 97 | 98.551 | ENSBTAG00000000423 | RBM4B | 96 | 98.780 | Bos_taurus |
| ENSG00000173933 | RBM4 | 100 | 97.253 | ENSBTAG00000031688 | RBM4 | 100 | 97.253 | Bos_taurus |
| ENSG00000173933 | RBM4 | 100 | 96.774 | ENSCJAG00000013666 | RBM4B | 100 | 96.774 | Callithrix_jacchus |
| ENSG00000173933 | RBM4 | 100 | 100.000 | ENSCAFG00000012388 | RBM4 | 100 | 99.178 | Canis_familiaris |
| ENSG00000173933 | RBM4 | 100 | 98.780 | ENSCAFG00000012378 | RBM4B | 96 | 98.780 | Canis_familiaris |
| ENSG00000173933 | RBM4 | 100 | 98.780 | ENSCAFG00020015519 | RBM4B | 96 | 98.780 | Canis_lupus_dingo |
| ENSG00000173933 | RBM4 | 100 | 100.000 | ENSCAFG00020015515 | RBM4 | 100 | 99.178 | Canis_lupus_dingo |
| ENSG00000173933 | RBM4 | 100 | 97.253 | ENSCHIG00000020613 | RBM4 | 100 | 97.253 | Capra_hircus |
| ENSG00000173933 | RBM4 | 97 | 98.551 | ENSCHIG00000008907 | RBM4B | 96 | 98.780 | Capra_hircus |
| ENSG00000173933 | RBM4 | 66 | 79.583 | ENSTSYG00000009580 | RBM4B | 96 | 79.668 | Carlito_syrichta |
| ENSG00000173933 | RBM4 | 94 | 87.850 | ENSTSYG00000036168 | - | 100 | 82.400 | Carlito_syrichta |
| ENSG00000173933 | RBM4 | 100 | 99.301 | ENSCAPG00000016600 | RBM4 | 100 | 99.301 | Cavia_aperea |
| ENSG00000173933 | RBM4 | 97 | 98.551 | ENSCAPG00000009570 | RBM4B | 96 | 98.780 | Cavia_aperea |
| ENSG00000173933 | RBM4 | 100 | 99.301 | ENSCPOG00000013194 | RBM4 | 100 | 99.301 | Cavia_porcellus |
| ENSG00000173933 | RBM4 | 97 | 98.551 | ENSCPOG00000013689 | RBM4B | 96 | 98.780 | Cavia_porcellus |
| ENSG00000173933 | RBM4 | 97 | 98.551 | ENSCCAG00000016501 | RBM4B | 98 | 97.857 | Cebus_capucinus |
| ENSG00000173933 | RBM4 | 99 | 72.535 | ENSCCAG00000023319 | - | 100 | 74.129 | Cebus_capucinus |
| ENSG00000173933 | RBM4 | 100 | 97.203 | ENSCCAG00000007508 | - | 100 | 97.203 | Cebus_capucinus |
| ENSG00000173933 | RBM4 | 97 | 98.551 | ENSCATG00000035353 | RBM4B | 98 | 97.857 | Cercocebus_atys |
| ENSG00000173933 | RBM4 | 100 | 99.301 | ENSCLAG00000014756 | RBM4 | 100 | 99.301 | Chinchilla_lanigera |
| ENSG00000173933 | RBM4 | 100 | 98.780 | ENSCLAG00000015287 | RBM4B | 96 | 98.780 | Chinchilla_lanigera |
| ENSG00000173933 | RBM4 | 97 | 98.551 | ENSCSAG00000008361 | RBM4B | 96 | 98.780 | Chlorocebus_sabaeus |
| ENSG00000173933 | RBM4 | 97 | 98.551 | ENSCHOG00000002892 | RBM4B | 96 | 98.780 | Choloepus_hoffmanni |
| ENSG00000173933 | RBM4 | 62 | 97.357 | ENSCHOG00000009753 | RBM14-RBM4 | 100 | 97.357 | Choloepus_hoffmanni |
| ENSG00000173933 | RBM4 | 100 | 96.341 | ENSCPBG00000023800 | - | 100 | 74.278 | Chrysemys_picta_bellii |
| ENSG00000173933 | RBM4 | 97 | 98.551 | ENSCANG00000021868 | RBM4B | 98 | 97.857 | Colobus_angolensis_palliatus |
| ENSG00000173933 | RBM4 | 100 | 98.601 | ENSCGRG00001016609 | - | 100 | 98.601 | Cricetulus_griseus_chok1gshd |
| ENSG00000173933 | RBM4 | 97 | 98.551 | ENSCGRG00001024693 | Rbm4b | 96 | 98.780 | Cricetulus_griseus_chok1gshd |
| ENSG00000173933 | RBM4 | 97 | 98.551 | ENSCGRG00000012112 | Rbm4b | 96 | 98.780 | Cricetulus_griseus_crigri |
| ENSG00000173933 | RBM4 | 100 | 98.601 | ENSCGRG00000010183 | - | 100 | 98.601 | Cricetulus_griseus_crigri |
| ENSG00000173933 | RBM4 | 97 | 98.551 | ENSDNOG00000019500 | - | 96 | 98.780 | Dasypus_novemcinctus |
| ENSG00000173933 | RBM4 | 100 | 99.390 | ENSDNOG00000032390 | RBM4 | 100 | 98.630 | Dasypus_novemcinctus |
| ENSG00000173933 | RBM4 | 100 | 97.561 | ENSDNOG00000045639 | - | 96 | 97.561 | Dasypus_novemcinctus |
| ENSG00000173933 | RBM4 | 97 | 98.551 | ENSDORG00000029108 | Rbm4b | 96 | 98.780 | Dipodomys_ordii |
| ENSG00000173933 | RBM4 | 100 | 98.171 | ENSDORG00000004538 | - | 100 | 97.902 | Dipodomys_ordii |
| ENSG00000173933 | RBM4 | 62 | 63.717 | ENSETEG00000018142 | - | 100 | 64.159 | Echinops_telfairi |
| ENSG00000173933 | RBM4 | 100 | 98.780 | ENSEASG00005005068 | RBM4B | 96 | 98.780 | Equus_asinus_asinus |
| ENSG00000173933 | RBM4 | 100 | 100.000 | ENSEASG00005005064 | RBM4 | 100 | 97.802 | Equus_asinus_asinus |
| ENSG00000173933 | RBM4 | 100 | 98.780 | ENSECAG00000022987 | RBM4B | 96 | 98.780 | Equus_caballus |
| ENSG00000173933 | RBM4 | 100 | 100.000 | ENSECAG00000022940 | RBM4 | 100 | 97.802 | Equus_caballus |
| ENSG00000173933 | RBM4 | 97 | 99.275 | ENSEEUG00000000058 | RBM4B | 96 | 99.390 | Erinaceus_europaeus |
| ENSG00000173933 | RBM4 | 97 | 98.551 | ENSFCAG00000007394 | RBM4B | 98 | 97.857 | Felis_catus |
| ENSG00000173933 | RBM4 | 100 | 100.000 | ENSFCAG00000029651 | RBM4 | 100 | 100.000 | Felis_catus |
| ENSG00000173933 | RBM4 | 97 | 97.101 | ENSFALG00000002073 | - | 100 | 68.108 | Ficedula_albicollis |
| ENSG00000173933 | RBM4 | 100 | 98.780 | ENSFDAG00000015530 | RBM4B | 96 | 98.780 | Fukomys_damarensis |
| ENSG00000173933 | RBM4 | 99 | 92.366 | ENSFDAG00000013661 | - | 96 | 92.366 | Fukomys_damarensis |
| ENSG00000173933 | RBM4 | 100 | 99.301 | ENSFDAG00000005447 | RBM4 | 100 | 99.301 | Fukomys_damarensis |
| ENSG00000173933 | RBM4 | 97 | 97.101 | ENSGALG00000037946 | - | 100 | 76.000 | Gallus_gallus |
| ENSG00000173933 | RBM4 | 97 | 97.101 | ENSGALG00000040489 | - | 86 | 95.745 | Gallus_gallus |
| ENSG00000173933 | RBM4 | 100 | 96.341 | ENSGAGG00000008553 | - | 100 | 74.607 | Gopherus_agassizii |
| ENSG00000173933 | RBM4 | 98 | 78.571 | ENSGGOG00000043023 | - | 98 | 79.562 | Gorilla_gorilla |
| ENSG00000173933 | RBM4 | 100 | 95.804 | ENSGGOG00000011698 | RBM4B | 98 | 97.857 | Gorilla_gorilla |
| ENSG00000173933 | RBM4 | 97 | 65.942 | ENSHGLG00000000117 | - | 95 | 66.027 | Heterocephalus_glaber_female |
| ENSG00000173933 | RBM4 | 100 | 85.976 | ENSHGLG00000002853 | - | 99 | 85.976 | Heterocephalus_glaber_female |
| ENSG00000173933 | RBM4 | 100 | 98.601 | ENSHGLG00000009991 | RBM4 | 100 | 98.601 | Heterocephalus_glaber_female |
| ENSG00000173933 | RBM4 | 100 | 98.780 | ENSHGLG00000016412 | - | 96 | 98.780 | Heterocephalus_glaber_female |
| ENSG00000173933 | RBM4 | 100 | 98.780 | ENSHGLG00100003978 | - | 96 | 98.780 | Heterocephalus_glaber_male |
| ENSG00000173933 | RBM4 | 100 | 98.601 | ENSHGLG00100003854 | RBM4 | 100 | 98.601 | Heterocephalus_glaber_male |
| ENSG00000173933 | RBM4 | 97 | 88.406 | ENSHGLG00100020033 | - | 100 | 85.976 | Heterocephalus_glaber_male |
| ENSG00000173933 | RBM4 | 97 | 98.551 | ENSSTOG00000011226 | RBM4B | 96 | 98.780 | Ictidomys_tridecemlineatus |
| ENSG00000173933 | RBM4 | 100 | 100.000 | ENSSTOG00000022811 | RBM4 | 100 | 100.000 | Ictidomys_tridecemlineatus |
| ENSG00000173933 | RBM4 | 93 | 75.188 | ENSJJAG00000003168 | - | 98 | 72.340 | Jaculus_jaculus |
| ENSG00000173933 | RBM4 | 100 | 94.512 | ENSJJAG00000020870 | Rbm4b | 96 | 94.512 | Jaculus_jaculus |
| ENSG00000173933 | RBM4 | 97 | 98.551 | ENSLAFG00000002285 | RBM4B | 96 | 98.780 | Loxodonta_africana |
| ENSG00000173933 | RBM4 | 97 | 98.551 | ENSLAFG00000028147 | - | 100 | 94.795 | Loxodonta_africana |
| ENSG00000173933 | RBM4 | 100 | 96.503 | ENSMFAG00000033927 | RBM4B | 97 | 98.551 | Macaca_fascicularis |
| ENSG00000173933 | RBM4 | 97 | 98.551 | ENSMNEG00000043371 | RBM4B | 98 | 97.857 | Macaca_nemestrina |
| ENSG00000173933 | RBM4 | 100 | 96.503 | ENSMLEG00000028563 | RBM4B | 98 | 97.857 | Mandrillus_leucophaeus |
| ENSG00000173933 | RBM4 | 97 | 98.551 | ENSMAUG00000017027 | Rbm4b | 96 | 98.780 | Mesocricetus_auratus |
| ENSG00000173933 | RBM4 | 100 | 97.902 | ENSMICG00000047975 | - | 99 | 97.902 | Microcebus_murinus |
| ENSG00000173933 | RBM4 | 100 | 95.804 | ENSMICG00000003817 | - | 100 | 95.804 | Microcebus_murinus |
| ENSG00000173933 | RBM4 | 100 | 98.780 | ENSMICG00000003820 | RBM4B | 98 | 97.857 | Microcebus_murinus |
| ENSG00000173933 | RBM4 | 97 | 98.551 | ENSMOCG00000001835 | Rbm4b | 96 | 98.780 | Microtus_ochrogaster |
| ENSG00000173933 | RBM4 | 100 | 96.951 | ENSMODG00000008056 | - | 100 | 90.489 | Monodelphis_domestica |
| ENSG00000173933 | RBM4 | 100 | 98.601 | MGP_CAROLIEiJ_G0022516 | - | 100 | 98.601 | Mus_caroli |
| ENSG00000173933 | RBM4 | 97 | 98.551 | MGP_CAROLIEiJ_G0022515 | Rbm4b | 96 | 98.780 | Mus_caroli |
| ENSG00000173933 | RBM4 | 99 | 87.013 | MGP_CAROLIEiJ_G0022514 | - | 100 | 71.038 | Mus_caroli |
| ENSG00000173933 | RBM4 | 100 | 95.890 | ENSMUSG00000096370 | Gm21992 | 100 | 95.890 | Mus_musculus |
| ENSG00000173933 | RBM4 | 100 | 95.804 | ENSMUSG00000033760 | Rbm4b | 96 | 98.780 | Mus_musculus |
| ENSG00000173933 | RBM4 | 100 | 98.601 | ENSMUSG00000094936 | Rbm4 | 100 | 98.837 | Mus_musculus |
| ENSG00000173933 | RBM4 | 97 | 98.551 | MGP_PahariEiJ_G0014012 | Rbm4b | 96 | 98.780 | Mus_pahari |
| ENSG00000173933 | RBM4 | 100 | 96.164 | MGP_PahariEiJ_G0014011 | - | 100 | 96.164 | Mus_pahari |
| ENSG00000173933 | RBM4 | 100 | 98.601 | MGP_PahariEiJ_G0014013 | - | 100 | 98.601 | Mus_pahari |
| ENSG00000173933 | RBM4 | 100 | 73.868 | MGP_SPRETEiJ_G0023429 | - | 100 | 75.694 | Mus_spretus |
| ENSG00000173933 | RBM4 | 100 | 87.013 | MGP_SPRETEiJ_G0023428 | - | 94 | 79.511 | Mus_spretus |
| ENSG00000173933 | RBM4 | 100 | 87.013 | MGP_SPRETEiJ_G0023430 | - | 100 | 79.511 | Mus_spretus |
| ENSG00000173933 | RBM4 | 97 | 98.551 | ENSMPUG00000003450 | RBM4B | 96 | 98.780 | Mustela_putorius_furo |
| ENSG00000173933 | RBM4 | 100 | 100.000 | ENSMPUG00000003443 | RBM4 | 100 | 98.904 | Mustela_putorius_furo |
| ENSG00000173933 | RBM4 | 100 | 99.390 | ENSMLUG00000004076 | RBM4 | 95 | 96.961 | Myotis_lucifugus |
| ENSG00000173933 | RBM4 | 100 | 98.780 | ENSMLUG00000001290 | RBM4B | 96 | 98.780 | Myotis_lucifugus |
| ENSG00000173933 | RBM4 | 97 | 98.551 | ENSNGAG00000016292 | Rbm4b | 96 | 98.780 | Nannospalax_galili |
| ENSG00000173933 | RBM4 | 100 | 100.000 | ENSNLEG00000035021 | RBM4 | 100 | 100.000 | Nomascus_leucogenys |
| ENSG00000173933 | RBM4 | 100 | 95.804 | ENSNLEG00000006213 | RBM4B | 98 | 97.857 | Nomascus_leucogenys |
| ENSG00000173933 | RBM4 | 100 | 98.171 | ENSMEUG00000001445 | - | 96 | 98.171 | Notamacropus_eugenii |
| ENSG00000173933 | RBM4 | 62 | 86.344 | ENSMEUG00000005739 | - | 100 | 86.404 | Notamacropus_eugenii |
| ENSG00000173933 | RBM4 | 100 | 98.780 | ENSOPRG00000009158 | RBM4B | 96 | 98.780 | Ochotona_princeps |
| ENSG00000173933 | RBM4 | 100 | 98.356 | ENSOPRG00000009149 | RBM4 | 100 | 98.356 | Ochotona_princeps |
| ENSG00000173933 | RBM4 | 100 | 95.804 | ENSODEG00000019143 | - | 100 | 95.804 | Octodon_degus |
| ENSG00000173933 | RBM4 | 97 | 99.275 | ENSODEG00000017696 | - | 96 | 99.390 | Octodon_degus |
| ENSG00000173933 | RBM4 | 100 | 85.366 | ENSODEG00000001866 | - | 96 | 85.366 | Octodon_degus |
| ENSG00000173933 | RBM4 | 100 | 97.203 | ENSODEG00000020039 | - | 100 | 97.203 | Octodon_degus |
| ENSG00000173933 | RBM4 | 100 | 76.923 | ENSODEG00000006635 | - | 100 | 76.923 | Octodon_degus |
| ENSG00000173933 | RBM4 | 100 | 98.780 | ENSOANG00000003143 | - | 81 | 98.780 | Ornithorhynchus_anatinus |
| ENSG00000173933 | RBM4 | 100 | 98.780 | ENSOANG00000011036 | - | 96 | 98.780 | Ornithorhynchus_anatinus |
| ENSG00000173933 | RBM4 | 100 | 99.452 | ENSOCUG00000004731 | RBM4 | 100 | 99.452 | Oryctolagus_cuniculus |
| ENSG00000173933 | RBM4 | 97 | 98.551 | ENSOCUG00000006695 | LARK | 96 | 98.780 | Oryctolagus_cuniculus |
| ENSG00000173933 | RBM4 | 97 | 98.551 | ENSOGAG00000015648 | RBM4B | 96 | 98.780 | Otolemur_garnettii |
| ENSG00000173933 | RBM4 | 100 | 99.452 | ENSOGAG00000015647 | RBM4 | 100 | 99.452 | Otolemur_garnettii |
| ENSG00000173933 | RBM4 | 97 | 98.551 | ENSOARG00000007759 | RBM4B | 96 | 98.780 | Ovis_aries |
| ENSG00000173933 | RBM4 | 97 | 100.000 | ENSOARG00000007700 | RBM4 | 96 | 100.000 | Ovis_aries |
| ENSG00000173933 | RBM4 | 100 | 95.804 | ENSPPAG00000015125 | RBM4B | 98 | 97.857 | Pan_paniscus |
| ENSG00000173933 | RBM4 | 97 | 98.551 | ENSPPRG00000003388 | RBM4B | 98 | 97.857 | Panthera_pardus |
| ENSG00000173933 | RBM4 | 100 | 100.000 | ENSPPRG00000003533 | RBM4 | 100 | 100.000 | Panthera_pardus |
| ENSG00000173933 | RBM4 | 97 | 98.551 | ENSPTIG00000015426 | RBM4B | 98 | 97.857 | Panthera_tigris_altaica |
| ENSG00000173933 | RBM4 | 100 | 99.301 | ENSPTIG00000016180 | RBM4 | 100 | 99.301 | Panthera_tigris_altaica |
| ENSG00000173933 | RBM4 | 100 | 95.804 | ENSPTRG00000003935 | RBM4B | 98 | 97.857 | Pan_troglodytes |
| ENSG00000173933 | RBM4 | 97 | 98.551 | ENSPANG00000020672 | RBM4B | 98 | 97.857 | Papio_anubis |
| ENSG00000173933 | RBM4 | 97 | 95.652 | ENSPSIG00000003477 | - | 95 | 77.740 | Pelodiscus_sinensis |
| ENSG00000173933 | RBM4 | 100 | 95.732 | ENSPSIG00000003430 | - | 96 | 95.732 | Pelodiscus_sinensis |
| ENSG00000173933 | RBM4 | 97 | 98.551 | ENSPEMG00000009144 | Rbm4b | 96 | 98.780 | Peromyscus_maniculatus_bairdii |
| ENSG00000173933 | RBM4 | 97 | 98.551 | ENSPPYG00000003010 | RBM4B | 96 | 98.780 | Pongo_abelii |
| ENSG00000173933 | RBM4 | 100 | 100.000 | ENSPPYG00000003012 | RBM4 | 100 | 98.901 | Pongo_abelii |
| ENSG00000173933 | RBM4 | 100 | 96.438 | ENSPCAG00000003393 | - | 100 | 96.438 | Procavia_capensis |
| ENSG00000173933 | RBM4 | 97 | 98.551 | ENSPCOG00000027885 | RBM4B | 99 | 98.561 | Propithecus_coquereli |
| ENSG00000173933 | RBM4 | 97 | 98.551 | ENSPVAG00000010487 | RBM4B | 96 | 98.780 | Pteropus_vampyrus |
| ENSG00000173933 | RBM4 | 100 | 100.000 | ENSPVAG00000010486 | RBM4 | 100 | 98.356 | Pteropus_vampyrus |
| ENSG00000173933 | RBM4 | 97 | 98.551 | ENSRNOG00000061795 | Rbm4b | 96 | 98.780 | Rattus_norvegicus |
| ENSG00000173933 | RBM4 | 100 | 98.601 | ENSRNOG00000050741 | Rbm4 | 100 | 98.601 | Rattus_norvegicus |
| ENSG00000173933 | RBM4 | 97 | 98.551 | ENSRBIG00000019394 | RBM4B | 98 | 97.857 | Rhinopithecus_bieti |
| ENSG00000173933 | RBM4 | 97 | 100.000 | ENSRROG00000032796 | - | 90 | 100.000 | Rhinopithecus_roxellana |
| ENSG00000173933 | RBM4 | 97 | 98.551 | ENSRROG00000032850 | RBM4B | 98 | 97.857 | Rhinopithecus_roxellana |
| ENSG00000173933 | RBM4 | 97 | 97.826 | ENSSBOG00000034474 | RBM4B | 98 | 97.143 | Saimiri_boliviensis_boliviensis |
| ENSG00000173933 | RBM4 | 100 | 98.780 | ENSSHAG00000008386 | - | 100 | 87.945 | Sarcophilus_harrisii |
| ENSG00000173933 | RBM4 | 97 | 98.551 | ENSSARG00000010710 | RBM4B | 96 | 98.780 | Sorex_araneus |
| ENSG00000173933 | RBM4 | 100 | 100.000 | ENSSSCG00000032437 | RBM4 | 100 | 99.178 | Sus_scrofa |
| ENSG00000173933 | RBM4 | 97 | 96.377 | ENSTGUG00000016779 | - | 94 | 77.740 | Taeniopygia_guttata |
| ENSG00000173933 | RBM4 | 96 | 100.000 | ENSTBEG00000003727 | - | 70 | 100.000 | Tupaia_belangeri |
| ENSG00000173933 | RBM4 | 100 | 98.780 | ENSTBEG00000003748 | RBM4B | 96 | 98.780 | Tupaia_belangeri |
| ENSG00000173933 | RBM4 | 100 | 97.260 | ENSTTRG00000011164 | RBM4 | 100 | 97.260 | Tursiops_truncatus |
| ENSG00000173933 | RBM4 | 100 | 98.171 | ENSTTRG00000011166 | RBM4B | 96 | 98.171 | Tursiops_truncatus |
| ENSG00000173933 | RBM4 | 100 | 100.000 | ENSUAMG00000014882 | RBM4 | 100 | 100.000 | Ursus_americanus |
| ENSG00000173933 | RBM4 | 100 | 98.780 | ENSUAMG00000004745 | RBM4B | 96 | 98.780 | Ursus_americanus |
| ENSG00000173933 | RBM4 | 100 | 99.301 | ENSUMAG00000011738 | - | 100 | 99.301 | Ursus_maritimus |
| ENSG00000173933 | RBM4 | 100 | 100.000 | ENSVPAG00000001949 | RBM4 | 100 | 96.712 | Vicugna_pacos |
| ENSG00000173933 | RBM4 | 97 | 97.826 | ENSVPAG00000001950 | RBM4B | 96 | 98.171 | Vicugna_pacos |
| ENSG00000173933 | RBM4 | 99 | 89.362 | ENSVVUG00000011717 | - | 100 | 85.714 | Vulpes_vulpes |
| ENSG00000173933 | RBM4 | 100 | 98.780 | ENSVVUG00000010619 | RBM4B | 96 | 98.780 | Vulpes_vulpes |
| ENSG00000173933 | RBM4 | 100 | 98.780 | ENSVVUG00000010622 | RBM4 | 100 | 98.630 | Vulpes_vulpes |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000381 | regulation of alternative mRNA splicing, via spliceosome | 21873635. | IBA | Process |
| GO:0000381 | regulation of alternative mRNA splicing, via spliceosome | 12628928.16260624.16777844.16934801.21518792. | IDA | Process |
| GO:0002190 | cap-independent translational initiation | 17284590. | IDA | Process |
| GO:0002192 | IRES-dependent translational initiation of linear mRNA | 17284590. | IDA | Process |
| GO:0003723 | RNA binding | 22658674.22681889. | HDA | Function |
| GO:0003723 | RNA binding | 21873635. | IBA | Function |
| GO:0003723 | RNA binding | 19561594. | IDA | Function |
| GO:0003729 | mRNA binding | 21518792. | IDA | Function |
| GO:0003730 | mRNA 3'-UTR binding | - | ISS | Function |
| GO:0005515 | protein binding | 12628928.16934801.17284590.19801630.21343338. | IPI | Function |
| GO:0005634 | nucleus | 21873635. | IBA | Component |
| GO:0005634 | nucleus | 16777844.16907643.17284590. | IDA | Component |
| GO:0005654 | nucleoplasm | 12628928. | IDA | Component |
| GO:0005730 | nucleolus | 12628928.16907643. | IDA | Component |
| GO:0005737 | cytoplasm | 12628928.16777844.16907643.17284590. | IDA | Component |
| GO:0005829 | cytosol | - | IDA | Component |
| GO:0006396 | RNA processing | 9169144. | TAS | Process |
| GO:0008270 | zinc ion binding | - | IEA | Function |
| GO:0010494 | cytoplasmic stress granule | 17284590. | IDA | Component |
| GO:0016607 | nuclear speck | 21873635. | IBA | Component |
| GO:0016607 | nuclear speck | 12628928.16907643. | IDA | Component |
| GO:0017148 | negative regulation of translation | 17284590. | IDA | Process |
| GO:0030154 | cell differentiation | - | IEA | Process |
| GO:0030332 | cyclin binding | 15159402. | IPI | Function |
| GO:0032055 | negative regulation of translation in response to stress | 17284590. | IDA | Process |
| GO:0035198 | miRNA binding | 19801630. | IDA | Function |
| GO:0035278 | miRNA mediated inhibition of translation | 19801630. | IDA | Process |
| GO:0043153 | entrainment of circadian clock by photoperiod | - | ISS | Process |
| GO:0045292 | mRNA cis splicing, via spliceosome | 21873635. | IBA | Process |
| GO:0045947 | negative regulation of translational initiation | 19801630. | IDA | Process |
| GO:0046685 | response to arsenic-containing substance | 17284590. | IDA | Process |
| GO:0046822 | regulation of nucleocytoplasmic transport | 12628928.17284590. | IDA | Process |
| GO:0051149 | positive regulation of muscle cell differentiation | 21518792. | IDA | Process |
| GO:0097157 | pre-mRNA intronic binding | 16260624. | IDA | Function |
| GO:0097158 | pre-mRNA intronic pyrimidine-rich binding | 16260624.16777844. | IDA | Function |
| GO:0097167 | circadian regulation of translation | - | ISS | Process |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CHOL | |||||||
| CHOL | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| DLBC | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRP | |||||||
| LAML | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SARC | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| TGCT | |||||||
| THCA | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| ESCA | |||
| TGCT |
| Cancer | P-value | Q-value |
|---|---|---|
| ACC | ||
| UCS | ||
| PRAD | ||
| PCPG | ||
| BLCA | ||
| READ | ||
| LAML | ||
| KICH | ||
| UCEC | ||
| GBM | ||
| LGG | ||
| THCA | ||
| UVM |