
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000546966 | CSRP2-202 | 754 | - | ENSP00000450056 | 193 (aa) | - | Q16527 |
| ENST00000547435 | CSRP2-203 | 632 | - | ENSP00000450143 | 187 (aa) | - | F8VQR7 |
| ENST00000547557 | CSRP2-204 | 628 | - | - | - (aa) | - | - |
| ENST00000552330 | CSRP2-207 | 910 | - | ENSP00000449824 | 243 (aa) | - | F8VW96 |
| ENST00000551725 | CSRP2-206 | 194 | - | - | - (aa) | - | - |
| ENST00000548783 | CSRP2-205 | 5078 | - | - | - (aa) | - | - |
| ENST00000311083 | CSRP2-201 | 943 | - | ENSP00000310901 | 193 (aa) | - | Q16527 |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000175183 | Asthma | 2E-6 | 27611488 |
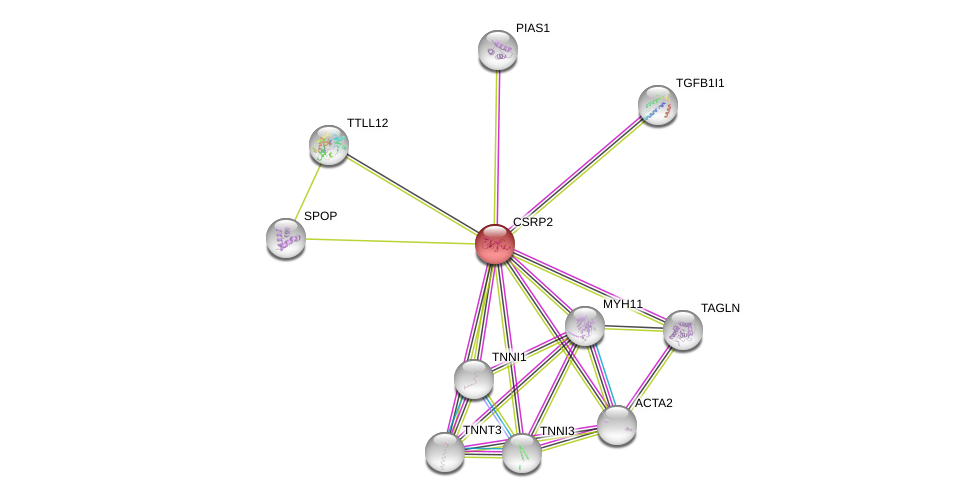
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000175183 | CSRP2 | 100 | 99.482 | ENSMUSG00000020186 | Csrp2 | 100 | 100.000 | Mus_musculus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0003674 | molecular_function | - | ND | Function |
| GO:0005515 | protein binding | 25416956. | IPI | Function |
| GO:0005634 | nucleus | 8626582. | NAS | Component |
| GO:0005925 | focal adhesion | 21423176. | HDA | Component |
| GO:0007275 | multicellular organism development | - | IEA | Process |
| GO:0030154 | cell differentiation | 8626582. | NAS | Process |
| GO:0046872 | metal ion binding | - | IEA | Function |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| GBM | |||||||
| KIRC | |||||||
| LIHC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| READ | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| ACC | |||
| DLBC | |||
| ESCA | |||
| HNSC | |||
| LUAD | |||
| PAAD | |||
| PRAD | |||
| READ | |||
| SKCM |
| Cancer | P-value | Q-value |
|---|---|---|
| STAD | ||
| MESO | ||
| ACC | ||
| HNSC | ||
| SKCM | ||
| PRAD | ||
| BRCA | ||
| KIRP | ||
| COAD | ||
| CESC | ||
| LAML | ||
| DLBC | ||
| LGG | ||
| LUAD | ||
| UVM |