
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000332444 | RRM_1 | PF00076.22 | 1.7e-24 | 1 | 1 |
| PID | Title | Article | Time | Author | Doi |
|---|---|---|---|---|---|
| 17029590 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000331173 | CSTF2T-201 | 4105 | - | ENSP00000332444 | 616 (aa) | - | Q9H0L4 |
| Pathway ID | Pathway Name | Source |
|---|---|---|
| hsa03015 | mRNA surveillance pathway | KEGG |
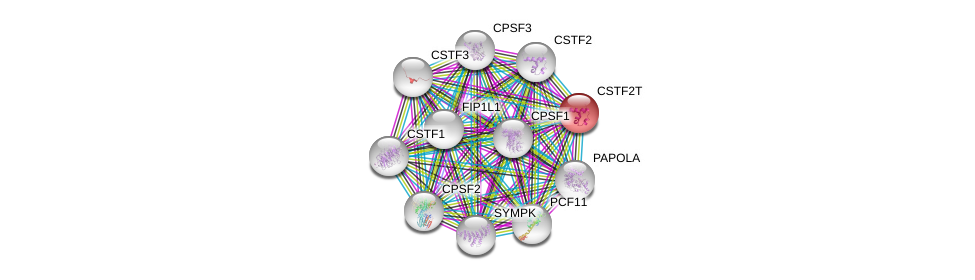
| Ensembl ID | Gene Symbol | Coverage | Identiy | Paralog | Gene Symbol | Coverage | Identiy |
|---|---|---|---|---|---|---|---|
| ENSG00000177613 | CSTF2T | 99 | 69.255 | ENSG00000101811 | CSTF2 | 84 | 88.360 |
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000177613 | CSTF2T | 100 | 91.396 | ENSAMEG00000015766 | CSTF2T | 100 | 91.396 | Ailuropoda_melanoleuca |
| ENSG00000177613 | CSTF2T | 54 | 85.843 | ENSANAG00000027508 | - | 89 | 85.843 | Aotus_nancymaae |
| ENSG00000177613 | CSTF2T | 100 | 88.144 | ENSBTAG00000018406 | CSTF2T | 100 | 88.612 | Bos_taurus |
| ENSG00000177613 | CSTF2T | 53 | 86.103 | ENSCJAG00000011554 | - | 97 | 86.103 | Callithrix_jacchus |
| ENSG00000177613 | CSTF2T | 53 | 80.243 | ENSCJAG00000039083 | - | 93 | 79.939 | Callithrix_jacchus |
| ENSG00000177613 | CSTF2T | 91 | 100.000 | ENSCAFG00000015567 | CSTF2T | 100 | 68.981 | Canis_familiaris |
| ENSG00000177613 | CSTF2T | 100 | 87.382 | ENSCHIG00000016590 | CSTF2T | 100 | 87.224 | Capra_hircus |
| ENSG00000177613 | CSTF2T | 100 | 92.903 | ENSTSYG00000007750 | CSTF2T | 100 | 93.226 | Carlito_syrichta |
| ENSG00000177613 | CSTF2T | 100 | 87.520 | ENSCPOG00000004195 | CSTF2T | 100 | 87.167 | Cavia_porcellus |
| ENSG00000177613 | CSTF2T | 54 | 85.629 | ENSCCAG00000020816 | - | 88 | 85.629 | Cebus_capucinus |
| ENSG00000177613 | CSTF2T | 53 | 86.486 | ENSCATG00000041868 | - | 95 | 86.486 | Cercocebus_atys |
| ENSG00000177613 | CSTF2T | 100 | 88.750 | ENSCLAG00000017916 | CSTF2T | 100 | 89.062 | Chinchilla_lanigera |
| ENSG00000177613 | CSTF2T | 100 | 97.258 | ENSCSAG00000018437 | CSTF2T | 100 | 97.258 | Chlorocebus_sabaeus |
| ENSG00000177613 | CSTF2T | 69 | 96.212 | ENSCHOG00000011888 | - | 68 | 96.212 | Choloepus_hoffmanni |
| ENSG00000177613 | CSTF2T | 53 | 85.758 | ENSCANG00000005843 | - | 92 | 85.758 | Colobus_angolensis_palliatus |
| ENSG00000177613 | CSTF2T | 100 | 85.826 | ENSCGRG00001003222 | Cstf2t | 100 | 85.202 | Cricetulus_griseus_chok1gshd |
| ENSG00000177613 | CSTF2T | 100 | 86.739 | ENSCGRG00000008682 | Cstf2t | 100 | 86.427 | Cricetulus_griseus_crigri |
| ENSG00000177613 | CSTF2T | 100 | 90.438 | ENSDNOG00000003283 | CSTF2T | 100 | 90.600 | Dasypus_novemcinctus |
| ENSG00000177613 | CSTF2T | 82 | 91.304 | ENSDORG00000011245 | Cstf2t | 100 | 87.893 | Dipodomys_ordii |
| ENSG00000177613 | CSTF2T | 100 | 92.432 | ENSEASG00005012929 | CSTF2T | 100 | 93.076 | Equus_asinus_asinus |
| ENSG00000177613 | CSTF2T | 100 | 91.214 | ENSECAG00000004701 | CSTF2T | 100 | 92.013 | Equus_caballus |
| ENSG00000177613 | CSTF2T | 90 | 81.579 | ENSELUG00000004079 | - | 98 | 57.346 | Esox_lucius |
| ENSG00000177613 | CSTF2T | 100 | 93.506 | ENSFCAG00000046300 | CSTF2T | 100 | 94.481 | Felis_catus |
| ENSG00000177613 | CSTF2T | 100 | 84.365 | ENSFDAG00000021170 | CSTF2T | 100 | 87.307 | Fukomys_damarensis |
| ENSG00000177613 | CSTF2T | 100 | 97.403 | ENSGGOG00000042388 | - | 100 | 97.403 | Gorilla_gorilla |
| ENSG00000177613 | CSTF2T | 100 | 97.403 | ENSGGOG00000036416 | - | 100 | 97.403 | Gorilla_gorilla |
| ENSG00000177613 | CSTF2T | 82 | 89.464 | ENSHGLG00000015478 | CSTF2T | 100 | 87.191 | Heterocephalus_glaber_female |
| ENSG00000177613 | CSTF2T | 82 | 89.464 | ENSHGLG00100000336 | CSTF2T | 100 | 87.211 | Heterocephalus_glaber_male |
| ENSG00000177613 | CSTF2T | 100 | 90.605 | ENSSTOG00000026303 | CSTF2T | 100 | 90.764 | Ictidomys_tridecemlineatus |
| ENSG00000177613 | CSTF2T | 100 | 84.253 | ENSLAFG00000002058 | CSTF2T | 100 | 85.227 | Loxodonta_africana |
| ENSG00000177613 | CSTF2T | 52 | 86.196 | ENSMFAG00000042009 | - | 97 | 86.196 | Macaca_fascicularis |
| ENSG00000177613 | CSTF2T | 54 | 85.928 | ENSMMUG00000042906 | - | 99 | 85.928 | Macaca_mulatta |
| ENSG00000177613 | CSTF2T | 52 | 86.196 | ENSMNEG00000031410 | - | 97 | 86.196 | Macaca_nemestrina |
| ENSG00000177613 | CSTF2T | 100 | 88.217 | ENSMAUG00000006359 | Cstf2t | 100 | 89.650 | Mesocricetus_auratus |
| ENSG00000177613 | CSTF2T | 100 | 87.402 | ENSMICG00000030551 | - | 100 | 89.928 | Microcebus_murinus |
| ENSG00000177613 | CSTF2T | 100 | 92.718 | ENSMICG00000000867 | - | 100 | 93.861 | Microcebus_murinus |
| ENSG00000177613 | CSTF2T | 100 | 86.250 | ENSMOCG00000020428 | Cstf2t | 100 | 86.875 | Microtus_ochrogaster |
| ENSG00000177613 | CSTF2T | 81 | 83.111 | ENSMALG00000008842 | - | 97 | 55.987 | Monopterus_albus |
| ENSG00000177613 | CSTF2T | 82 | 89.749 | MGP_CAROLIEiJ_G0022872 | Cstf2t | 100 | 88.766 | Mus_caroli |
| ENSG00000177613 | CSTF2T | 82 | 89.168 | ENSMUSG00000053536 | Cstf2t | 100 | 87.207 | Mus_musculus |
| ENSG00000177613 | CSTF2T | 82 | 88.168 | MGP_PahariEiJ_G0014365 | Cstf2t | 100 | 85.957 | Mus_pahari |
| ENSG00000177613 | CSTF2T | 100 | 83.645 | MGP_SPRETEiJ_G0023790 | Cstf2t | 100 | 87.676 | Mus_spretus |
| ENSG00000177613 | CSTF2T | 100 | 91.613 | ENSMPUG00000020141 | CSTF2T | 100 | 91.613 | Mustela_putorius_furo |
| ENSG00000177613 | CSTF2T | 100 | 86.159 | ENSMLUG00000011187 | - | 100 | 86.314 | Myotis_lucifugus |
| ENSG00000177613 | CSTF2T | 100 | 83.226 | ENSMLUG00000027140 | - | 100 | 83.226 | Myotis_lucifugus |
| ENSG00000177613 | CSTF2T | 100 | 89.172 | ENSNGAG00000003304 | Cstf2t | 100 | 89.490 | Nannospalax_galili |
| ENSG00000177613 | CSTF2T | 100 | 94.042 | ENSNLEG00000012025 | CSTF2T | 100 | 94.042 | Nomascus_leucogenys |
| ENSG00000177613 | CSTF2T | 100 | 84.062 | ENSODEG00000012386 | CSTF2T | 100 | 83.906 | Octodon_degus |
| ENSG00000177613 | CSTF2T | 100 | 83.117 | ENSOCUG00000004647 | - | 100 | 83.117 | Oryctolagus_cuniculus |
| ENSG00000177613 | CSTF2T | 100 | 93.831 | ENSOGAG00000014565 | CSTF2T | 100 | 93.994 | Otolemur_garnettii |
| ENSG00000177613 | CSTF2T | 100 | 88.608 | ENSOARG00000019124 | - | 100 | 88.924 | Ovis_aries |
| ENSG00000177613 | CSTF2T | 100 | 87.520 | ENSOARG00000013688 | - | 100 | 88.144 | Ovis_aries |
| ENSG00000177613 | CSTF2T | 53 | 86.280 | ENSPPAG00000032242 | - | 96 | 86.280 | Pan_paniscus |
| ENSG00000177613 | CSTF2T | 100 | 93.831 | ENSPPRG00000018659 | CSTF2T | 100 | 94.156 | Panthera_pardus |
| ENSG00000177613 | CSTF2T | 100 | 90.584 | ENSPTIG00000021145 | CSTF2T | 100 | 91.396 | Panthera_tigris_altaica |
| ENSG00000177613 | CSTF2T | 53 | 86.280 | ENSPTRG00000043097 | - | 96 | 86.280 | Pan_troglodytes |
| ENSG00000177613 | CSTF2T | 52 | 86.196 | ENSPANG00000029734 | - | 97 | 86.196 | Papio_anubis |
| ENSG00000177613 | CSTF2T | 98 | 52.019 | ENSPFOG00000016756 | CSTF2T | 94 | 66.379 | Poecilia_formosa |
| ENSG00000177613 | CSTF2T | 100 | 96.640 | ENSPPYG00000002441 | CSTF2T | 100 | 96.640 | Pongo_abelii |
| ENSG00000177613 | CSTF2T | 100 | 90.468 | ENSPCAG00000000046 | CSTF2T | 100 | 91.599 | Procavia_capensis |
| ENSG00000177613 | CSTF2T | 99 | 92.683 | ENSPCOG00000010982 | CSTF2T | 99 | 93.669 | Propithecus_coquereli |
| ENSG00000177613 | CSTF2T | 100 | 92.370 | ENSPVAG00000006274 | CSTF2T | 100 | 92.370 | Pteropus_vampyrus |
| ENSG00000177613 | CSTF2T | 100 | 88.013 | ENSRNOG00000050289 | Cstf2t | 100 | 88.013 | Rattus_norvegicus |
| ENSG00000177613 | CSTF2T | 100 | 97.727 | ENSRBIG00000029324 | CSTF2T | 100 | 97.727 | Rhinopithecus_bieti |
| ENSG00000177613 | CSTF2T | 100 | 97.890 | ENSRROG00000042507 | CSTF2T | 100 | 97.890 | Rhinopithecus_roxellana |
| ENSG00000177613 | CSTF2T | 100 | 96.916 | ENSSBOG00000022638 | CSTF2T | 100 | 96.916 | Saimiri_boliviensis_boliviensis |
| ENSG00000177613 | CSTF2T | 88 | 95.276 | ENSSARG00000002013 | - | 87 | 94.094 | Sorex_araneus |
| ENSG00000177613 | CSTF2T | 79 | 90.816 | ENSSPAG00000009247 | - | 96 | 54.984 | Stegastes_partitus |
| ENSG00000177613 | CSTF2T | 100 | 93.831 | ENSSSCG00000010430 | CSTF2T | 100 | 93.831 | Sus_scrofa |
| ENSG00000177613 | CSTF2T | 99 | 52.572 | ENSTRUG00000004323 | - | 97 | 51.713 | Takifugu_rubripes |
| ENSG00000177613 | CSTF2T | 100 | 82.792 | ENSTBEG00000014379 | CSTF2T | 100 | 82.792 | Tupaia_belangeri |
| ENSG00000177613 | CSTF2T | 100 | 92.532 | ENSTTRG00000003341 | CSTF2T | 100 | 92.857 | Tursiops_truncatus |
| ENSG00000177613 | CSTF2T | 100 | 88.612 | ENSUMAG00000011141 | CSTF2T | 100 | 90.016 | Ursus_maritimus |
| ENSG00000177613 | CSTF2T | 65 | 96.020 | ENSVVUG00000002575 | CSTF2T | 96 | 95.631 | Vulpes_vulpes |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000398 | mRNA splicing, via spliceosome | - | TAS | Process |
| GO:0003723 | RNA binding | 22658674.22681889. | HDA | Function |
| GO:0003723 | RNA binding | 21873635. | IBA | Function |
| GO:0003729 | mRNA binding | 21873635. | IBA | Function |
| GO:0005515 | protein binding | 16189514.25416956. | IPI | Function |
| GO:0005654 | nucleoplasm | - | IDA | Component |
| GO:0005654 | nucleoplasm | - | TAS | Component |
| GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex | 21873635. | IBA | Component |
| GO:0006369 | termination of RNA polymerase II transcription | - | TAS | Process |
| GO:0031124 | mRNA 3'-end processing | - | TAS | Process |
| GO:0098789 | pre-mRNA cleavage required for polyadenylation | 21873635. | IBA | Process |