
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000431620 | PARP | PF00644.20 | 1.2e-26 | 1 | 1 |
| ENSP00000325618 | PARP | PF00644.20 | 1.2e-26 | 1 | 1 |
| ENSP00000434776 | PARP | PF00644.20 | 1.3e-26 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000313059 | PARP10-202 | 1090 | - | ENSP00000314320 | 339 (aa) | - | F8W8G7 |
| ENST00000531537 | PARP10-218 | 578 | - | ENSP00000432204 | 144 (aa) | - | E9PIA6 |
| ENST00000528914 | PARP10-213 | 589 | - | ENSP00000434005 | 135 (aa) | - | E9PSE7 |
| ENST00000529311 | PARP10-215 | 529 | - | ENSP00000434411 | 106 (aa) | - | E9PQQ6 |
| ENST00000526007 | PARP10-207 | 2179 | - | - | - (aa) | - | - |
| ENST00000525486 | PARP10-204 | 564 | - | ENSP00000435793 | 146 (aa) | - | E9PIK9 |
| ENST00000529842 | PARP10-216 | 696 | - | ENSP00000436697 | 144 (aa) | - | E9PIA6 |
| ENST00000528580 | PARP10-211 | 488 | - | - | - (aa) | - | - |
| ENST00000525773 | PARP10-205 | 3404 | - | ENSP00000434776 | 1037 (aa) | - | E9PNI7 |
| ENST00000532311 | PARP10-220 | 481 | - | ENSP00000435161 | 83 (aa) | - | E9PLE8 |
| ENST00000528963 | PARP10-214 | 581 | - | - | - (aa) | - | - |
| ENST00000313028 | PARP10-201 | 3497 | - | ENSP00000325618 | 1025 (aa) | - | Q53GL7 |
| ENST00000532660 | PARP10-221 | 545 | - | - | - (aa) | - | - |
| ENST00000527262 | PARP10-209 | 3027 | - | ENSP00000432733 | 764 (aa) | - | E9PPE7 |
| ENST00000533665 | PARP10-222 | 935 | - | - | - (aa) | - | - |
| ENST00000528625 | PARP10-212 | 685 | - | ENSP00000431818 | 172 (aa) | - | E9PJI2 |
| ENST00000524918 | PARP10-203 | 3469 | XM_011517336 | ENSP00000431620 | 1016 (aa) | XP_011515638 | E9PK67 |
| ENST00000525879 | PARP10-206 | 939 | - | ENSP00000436301 | 178 (aa) | - | E9PPV8 |
| ENST00000528136 | PARP10-210 | 899 | - | ENSP00000431250 | 128 (aa) | - | E9PM86 |
| ENST00000526985 | PARP10-208 | 571 | - | - | - (aa) | - | - |
| ENST00000530478 | PARP10-217 | 588 | - | ENSP00000436333 | 88 (aa) | - | E9PPU2 |
| ENST00000531707 | PARP10-219 | 500 | - | ENSP00000433990 | 81 (aa) | - | E9PSG8 |
| ENST00000534737 | PARP10-223 | 550 | - | - | - (aa) | - | - |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000178685 | rs11784833 | 8 | 143989244 | C | LDL cholesterol | 25961943 | [0.028-0.056] s.d. increase | 0.042 | EFO_0004611 |
| ENSG00000178685 | rs6985603 | 8 | 144012260 | ? | Tonsillectomy | 28928442 | [0.023-0.056] unit increase | 0.0398 | EFO_0007924 |
| ENSG00000178685 | rs56261297 | 8 | 143992685 | ? | Mean corpuscular volume | 29403010 | [0.022-0.045] unit increase novel | 0.03373 | EFO_0004526 |
| ENSG00000178685 | rs6420181 | 8 | 144007080 | ? | Mean corpuscular hemoglobin | 29403010 | [0.023-0.045] unit increase novel | 0.0336 | EFO_0004527 |
| ENSG00000178685 | rs11136252 | 8 | 144010325 | ? | Fibrinogen levels | 28107422 | unit increase | 0.0056 | EFO_0004623 |
| ENSG00000178685 | rs11784833 | 8 | 143989244 | C | Red cell distribution width | 28957414 | [0.015-0.031] unit decrease | 0.0231467 | EFO_0005192 |
| ENSG00000178685 | rs11784833 | 8 | 143989244 | T | Low density lipoprotein cholesterol levels | 29507422 | unit decrease | 0.025 | EFO_0004611 |
| ENSG00000178685 | rs11784833 | 8 | 143989244 | T | Low density lipoprotein cholesterol levels | 29507422 | unit decrease | 0.027 | EFO_0004611 |
| ENSG00000178685 | rs11784833 | 8 | 143989244 | T | Total cholesterol levels | 29507422 | unit decrease | 0.022 | EFO_0004574 |
| ENSG00000178685 | rs11784833 | 8 | 143989244 | ? | Red cell distribution width | 30595370 | EFO_0005192 |
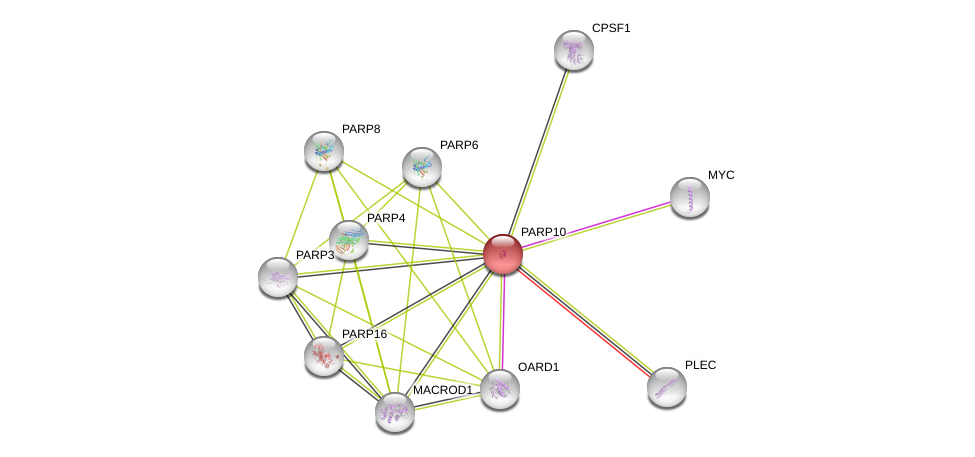
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000178685 | PARP10 | 86 | 54.167 | ENSAPOG00000016910 | si:ch1073-296d18.1 | 65 | 46.067 | Acanthochromis_polyacanthus |
| ENSG00000178685 | PARP10 | 55 | 33.333 | ENSAOCG00000007023 | - | 70 | 32.865 | Amphiprion_ocellaris |
| ENSG00000178685 | PARP10 | 99 | 94.776 | ENSANAG00000037218 | PARP10 | 99 | 84.195 | Aotus_nancymaae |
| ENSG00000178685 | PARP10 | 97 | 80.851 | ENSBTAG00000009677 | PARP10 | 99 | 65.982 | Bos_taurus |
| ENSG00000178685 | PARP10 | 99 | 94.488 | ENSCJAG00000034531 | PARP10 | 100 | 85.274 | Callithrix_jacchus |
| ENSG00000178685 | PARP10 | 100 | 80.822 | ENSCHIG00000008072 | PARP10 | 100 | 66.569 | Capra_hircus |
| ENSG00000178685 | PARP10 | 94 | 88.148 | ENSTSYG00000005439 | PARP10 | 99 | 75.588 | Carlito_syrichta |
| ENSG00000178685 | PARP10 | 98 | 80.303 | ENSCAPG00000014295 | PARP10 | 99 | 73.810 | Cavia_aperea |
| ENSG00000178685 | PARP10 | 98 | 80.303 | ENSCPOG00000014235 | PARP10 | 100 | 66.371 | Cavia_porcellus |
| ENSG00000178685 | PARP10 | 100 | 91.573 | ENSCCAG00000024610 | PARP10 | 100 | 84.957 | Cebus_capucinus |
| ENSG00000178685 | PARP10 | 100 | 97.917 | ENSCATG00000025822 | PARP10 | 100 | 92.683 | Cercocebus_atys |
| ENSG00000178685 | PARP10 | 97 | 83.495 | ENSCLAG00000013744 | PARP10 | 100 | 67.777 | Chinchilla_lanigera |
| ENSG00000178685 | PARP10 | 98 | 97.727 | ENSCSAG00000008843 | PARP10 | 100 | 92.857 | Chlorocebus_sabaeus |
| ENSG00000178685 | PARP10 | 72 | 56.284 | ENSCPBG00000004075 | PARP10 | 85 | 56.284 | Chrysemys_picta_bellii |
| ENSG00000178685 | PARP10 | 100 | 97.222 | ENSCANG00000043564 | PARP10 | 97 | 90.723 | Colobus_angolensis_palliatus |
| ENSG00000178685 | PARP10 | 97 | 76.471 | ENSCGRG00001012953 | Parp10 | 100 | 64.356 | Cricetulus_griseus_chok1gshd |
| ENSG00000178685 | PARP10 | 97 | 76.471 | ENSCGRG00000003088 | Parp10 | 100 | 64.356 | Cricetulus_griseus_crigri |
| ENSG00000178685 | PARP10 | 73 | 53.297 | ENSDARG00000087145 | si:ch1073-296d18.1 | 68 | 53.297 | Danio_rerio |
| ENSG00000178685 | PARP10 | 90 | 78.788 | ENSDNOG00000046110 | PARP10 | 99 | 64.050 | Dasypus_novemcinctus |
| ENSG00000178685 | PARP10 | 99 | 75.238 | ENSETEG00000014689 | - | 56 | 72.000 | Echinops_telfairi |
| ENSG00000178685 | PARP10 | 89 | 84.043 | ENSEASG00005009729 | PARP10 | 99 | 71.775 | Equus_asinus_asinus |
| ENSG00000178685 | PARP10 | 97 | 75.573 | ENSEEUG00000005507 | PARP10 | 91 | 73.826 | Erinaceus_europaeus |
| ENSG00000178685 | PARP10 | 100 | 50.000 | ENSELUG00000001455 | si:ch1073-296d18.1 | 70 | 52.660 | Esox_lucius |
| ENSG00000178685 | PARP10 | 98 | 84.091 | ENSFDAG00000015949 | PARP10 | 100 | 66.272 | Fukomys_damarensis |
| ENSG00000178685 | PARP10 | 90 | 44.211 | ENSGMOG00000000853 | si:ch1073-296d18.1 | 80 | 33.043 | Gadus_morhua |
| ENSG00000178685 | PARP10 | 94 | 50.633 | ENSGACG00000008798 | si:ch1073-296d18.1 | 84 | 33.506 | Gasterosteus_aculeatus |
| ENSG00000178685 | PARP10 | 100 | 99.438 | ENSGGOG00000001802 | PARP10 | 100 | 98.049 | Gorilla_gorilla |
| ENSG00000178685 | PARP10 | 99 | 82.090 | ENSHGLG00000010423 | PARP10 | 96 | 66.978 | Heterocephalus_glaber_female |
| ENSG00000178685 | PARP10 | 84 | 51.913 | ENSIPUG00000011187 | parp10 | 74 | 51.913 | Ictalurus_punctatus |
| ENSG00000178685 | PARP10 | 97 | 84.706 | ENSJJAG00000017801 | Parp10 | 99 | 64.243 | Jaculus_jaculus |
| ENSG00000178685 | PARP10 | 55 | 33.275 | ENSLBEG00000003798 | si:ch1073-296d18.1 | 90 | 33.042 | Labrus_bergylta |
| ENSG00000178685 | PARP10 | 99 | 52.874 | ENSLACG00000008645 | PARP10 | 94 | 38.435 | Latimeria_chalumnae |
| ENSG00000178685 | PARP10 | 85 | 52.174 | ENSLOCG00000008505 | si:ch1073-296d18.1 | 80 | 35.017 | Lepisosteus_oculatus |
| ENSG00000178685 | PARP10 | 96 | 63.546 | ENSLAFG00000004191 | PARP10 | 98 | 63.817 | Loxodonta_africana |
| ENSG00000178685 | PARP10 | 99 | 98.851 | ENSMFAG00000044942 | PARP10 | 100 | 92.759 | Macaca_fascicularis |
| ENSG00000178685 | PARP10 | 99 | 98.851 | ENSMMUG00000000449 | PARP10 | 100 | 92.671 | Macaca_mulatta |
| ENSG00000178685 | PARP10 | 99 | 98.851 | ENSMNEG00000034827 | PARP10 | 100 | 92.857 | Macaca_nemestrina |
| ENSG00000178685 | PARP10 | 100 | 97.222 | ENSMLEG00000024662 | PARP10 | 100 | 92.421 | Mandrillus_leucophaeus |
| ENSG00000178685 | PARP10 | 99 | 88.060 | ENSMICG00000016227 | PARP10 | 100 | 76.774 | Microcebus_murinus |
| ENSG00000178685 | PARP10 | 97 | 77.647 | ENSMOCG00000000179 | Parp10 | 100 | 64.229 | Microtus_ochrogaster |
| ENSG00000178685 | PARP10 | 98 | 66.667 | ENSMODG00000010393 | - | 86 | 63.600 | Monodelphis_domestica |
| ENSG00000178685 | PARP10 | 97 | 78.824 | MGP_CAROLIEiJ_G0019914 | Parp10 | 100 | 78.182 | Mus_caroli |
| ENSG00000178685 | PARP10 | 97 | 78.824 | ENSMUSG00000063268 | Parp10 | 99 | 78.182 | Mus_musculus |
| ENSG00000178685 | PARP10 | 97 | 80.000 | MGP_PahariEiJ_G0019924 | Parp10 | 100 | 77.273 | Mus_pahari |
| ENSG00000178685 | PARP10 | 97 | 78.824 | MGP_SPRETEiJ_G0020817 | Parp10 | 100 | 77.273 | Mus_spretus |
| ENSG00000178685 | PARP10 | 79 | 66.991 | ENSMLUG00000002730 | PARP10 | 100 | 67.275 | Myotis_lucifugus |
| ENSG00000178685 | PARP10 | 98 | 83.688 | ENSNGAG00000023968 | Parp10 | 100 | 67.682 | Nannospalax_galili |
| ENSG00000178685 | PARP10 | 99 | 98.305 | ENSNLEG00000002423 | PARP10 | 100 | 94.118 | Nomascus_leucogenys |
| ENSG00000178685 | PARP10 | 95 | 66.197 | ENSMEUG00000001906 | - | 92 | 66.452 | Notamacropus_eugenii |
| ENSG00000178685 | PARP10 | 98 | 71.970 | ENSOPRG00000015616 | - | 66 | 71.739 | Ochotona_princeps |
| ENSG00000178685 | PARP10 | 98 | 75.887 | ENSODEG00000014276 | PARP10 | 100 | 64.706 | Octodon_degus |
| ENSG00000178685 | PARP10 | 86 | 50.000 | ENSONIG00000007669 | si:ch1073-296d18.1 | 74 | 39.835 | Oreochromis_niloticus |
| ENSG00000178685 | PARP10 | 92 | 55.446 | ENSOANG00000013123 | - | 94 | 52.529 | Ornithorhynchus_anatinus |
| ENSG00000178685 | PARP10 | 99 | 80.303 | ENSOGAG00000033747 | PARP10 | 99 | 73.608 | Otolemur_garnettii |
| ENSG00000178685 | PARP10 | 99 | 80.000 | ENSOARG00000015581 | PARP10 | 99 | 65.962 | Ovis_aries |
| ENSG00000178685 | PARP10 | 100 | 99.438 | ENSPPAG00000041157 | PARP10 | 100 | 98.168 | Pan_paniscus |
| ENSG00000178685 | PARP10 | 100 | 99.438 | ENSPTRG00000043606 | PARP10 | 100 | 98.049 | Pan_troglodytes |
| ENSG00000178685 | PARP10 | 99 | 98.851 | ENSPANG00000023348 | PARP10 | 100 | 92.768 | Papio_anubis |
| ENSG00000178685 | PARP10 | 92 | 44.330 | ENSPFOG00000013067 | si:ch1073-296d18.1 | 100 | 30.324 | Poecilia_formosa |
| ENSG00000178685 | PARP10 | 55 | 33.108 | ENSPLAG00000022710 | - | 78 | 32.487 | Poecilia_latipinna |
| ENSG00000178685 | PARP10 | 55 | 32.982 | ENSPMEG00000019504 | - | 77 | 32.632 | Poecilia_mexicana |
| ENSG00000178685 | PARP10 | 100 | 100.000 | ENSPPYG00000018955 | PARP10 | 100 | 91.033 | Pongo_abelii |
| ENSG00000178685 | PARP10 | 95 | 82.143 | ENSPCAG00000001115 | PARP10 | 90 | 78.231 | Procavia_capensis |
| ENSG00000178685 | PARP10 | 100 | 88.194 | ENSPCOG00000028616 | PARP10 | 100 | 77.171 | Propithecus_coquereli |
| ENSG00000178685 | PARP10 | 91 | 83.333 | ENSPVAG00000006369 | PARP10 | 99 | 68.457 | Pteropus_vampyrus |
| ENSG00000178685 | PARP10 | 99 | 44.286 | ENSPNAG00000024484 | si:ch1073-296d18.1 | 65 | 51.075 | Pygocentrus_nattereri |
| ENSG00000178685 | PARP10 | 98 | 73.759 | ENSRNOG00000004361 | Parp10 | 100 | 64.138 | Rattus_norvegicus |
| ENSG00000178685 | PARP10 | 99 | 96.528 | ENSRBIG00000040506 | PARP10 | 100 | 92.478 | Rhinopithecus_bieti |
| ENSG00000178685 | PARP10 | 98 | 97.163 | ENSRROG00000039086 | PARP10 | 100 | 90.944 | Rhinopithecus_roxellana |
| ENSG00000178685 | PARP10 | 98 | 93.939 | ENSSBOG00000008301 | PARP10 | 100 | 82.583 | Saimiri_boliviensis_boliviensis |
| ENSG00000178685 | PARP10 | 98 | 63.121 | ENSSHAG00000002568 | - | 87 | 60.956 | Sarcophilus_harrisii |
| ENSG00000178685 | PARP10 | 55 | 33.275 | ENSSLDG00000003545 | - | 79 | 33.392 | Seriola_lalandi_dorsalis |
| ENSG00000178685 | PARP10 | 99 | 48.611 | ENSTNIG00000014023 | si:ch1073-296d18.1 | 85 | 33.846 | Tetraodon_nigroviridis |
| ENSG00000178685 | PARP10 | 100 | 78.409 | ENSTTRG00000006314 | PARP10 | 99 | 68.462 | Tursiops_truncatus |
| ENSG00000178685 | PARP10 | 55 | 33.000 | ENSXCOG00000003615 | - | 78 | 32.877 | Xiphophorus_couchianus |
| ENSG00000178685 | PARP10 | 55 | 33.275 | ENSXMAG00000025112 | - | 77 | 33.275 | Xiphophorus_maculatus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0003950 | NAD+ ADP-ribosyltransferase activity | 23473667.23575687. | IMP | Function |
| GO:0005515 | protein binding | 15674325.22176891.23473667.23481255.24695737. | IPI | Function |
| GO:0005634 | nucleus | 15674325.22176891.23473667. | IDA | Component |
| GO:0005730 | nucleolus | - | IDA | Component |
| GO:0005737 | cytoplasm | 15674325.22176891.23473667. | IDA | Component |
| GO:0005829 | cytosol | - | IDA | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0006471 | protein ADP-ribosylation | 23575687. | IMP | Process |
| GO:0010629 | negative regulation of gene expression | 23575687. | IMP | Process |
| GO:0010847 | regulation of chromatin assembly | 15674325. | IDA | Process |
| GO:0019985 | translesion synthesis | 24695737. | IDA | Process |
| GO:0032088 | negative regulation of NF-kappaB transcription factor activity | 23575687. | IMP | Process |
| GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway | - | TAS | Process |
| GO:0043231 | intracellular membrane-bounded organelle | - | IDA | Component |
| GO:0045071 | negative regulation of viral genome replication | 22176891. | IMP | Process |
| GO:0048147 | negative regulation of fibroblast proliferation | 15674325. | IDA | Process |
| GO:0070212 | protein poly-ADP-ribosylation | 15674325. | IDA | Process |
| GO:0070212 | protein poly-ADP-ribosylation | 23473667. | IMP | Process |
| GO:0070213 | protein auto-ADP-ribosylation | 18851833.25043379. | IDA | Process |
| GO:0070213 | protein auto-ADP-ribosylation | 23575687. | IMP | Process |
| GO:0070530 | K63-linked polyubiquitin modification-dependent protein binding | 23575687. | IMP | Function |
| GO:1900045 | negative regulation of protein K63-linked ubiquitination | 23575687. | IMP | Process |
| GO:1990404 | protein ADP-ribosylase activity | 18851833.25043379. | IDA | Function |
| GO:1990404 | protein ADP-ribosylase activity | 24695737. | IMP | Function |