
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000584044 | PFAS-209 | 557 | - | ENSP00000464689 | 135 (aa) | - | J3QSH6 |
| ENST00000546020 | PFAS-202 | 2652 | - | ENSP00000443244 | 385 (aa) | - | H0YGH1 |
| ENST00000625942 | PFAS-212 | 396 | - | ENSP00000487260 | 131 (aa) | - | J3QSG0 |
| ENST00000580251 | PFAS-204 | 559 | - | - | - (aa) | - | - |
| ENST00000578979 | PFAS-203 | 765 | - | ENSP00000463103 | 62 (aa) | - | J3KTQ5 |
| ENST00000581288 | PFAS-207 | 527 | - | - | - (aa) | - | - |
| ENST00000585183 | PFAS-210 | 581 | - | ENSP00000463362 | 87 (aa) | - | J3QL39 |
| ENST00000585319 | PFAS-211 | 598 | - | - | - (aa) | - | - |
| ENST00000583059 | PFAS-208 | 349 | - | ENSP00000462874 | 116 (aa) | - | J3KT98 |
| ENST00000580356 | PFAS-205 | 4242 | - | ENSP00000464671 | 131 (aa) | - | J3QSG0 |
| ENST00000581242 | PFAS-206 | 553 | - | ENSP00000463045 | 103 (aa) | - | J3KTL4 |
| ENST00000314666 | PFAS-201 | 5371 | XM_006721546 | ENSP00000313490 | 1338 (aa) | XP_006721609 | O15067 |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000178921 | Cholesterol, LDL | 1E-8 | 26780889 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000178921 | rs4791641 | 17 | 8257831 | T | Red blood cell count | 27863252 | [0.016-0.03] unit decrease | 0.02332721 | EFO_0004305 |
| ENSG00000178921 | rs4791641 | 17 | 8257831 | C | LDL cholesterol | 26780889 | [0.013-0.029] unit increase | 0.021 | EFO_0004611 |
| ENSG00000178921 | rs56309584 | 17 | 8261034 | C | Initial pursuit acceleration | 29064472 | EFO_0008434 | ||
| ENSG00000178921 | rs78296773 | 17 | 8264644 | G | Self-reported math ability | 30038396 | [0.013-0.022] unit decrease | 0.0179 | EFO_0004875 |
| ENSG00000178921 | rs78296773 | 17 | 8264644 | T | Self-reported math ability (MTAG) | 30038396 | [0.013-0.021] unit increase | 0.0167 | EFO_0004875 |
| ENSG00000178921 | rs4791641 | 17 | 8257831 | T | Hematocrit | 27863252 | [0.023-0.036] unit decrease | 0.02955392 | EFO_0004348 |
| ENSG00000178921 | rs4791641 | 17 | 8257831 | T | Hemoglobin concentration | 27863252 | [0.016-0.03] unit decrease | 0.02285043 | EFO_0004509 |
| ENSG00000178921 | rs10468482 | 17 | 8259502 | C | Mean corpuscular hemoglobin concentration | 27863252 | [0.017-0.034] unit increase | 0.0253879 | EFO_0004528 |
| ENSG00000178921 | rs4791641 | 17 | 8257831 | T | LDL cholesterol levels | 28334899 | [0.013-0.027] unit decrease (EA Beta value) | 0.0202 | EFO_0004611 |
| ENSG00000178921 | rs9891699 | 17 | 8253992 | ? | Red blood cell count | 30595370 | EFO_0004305 |
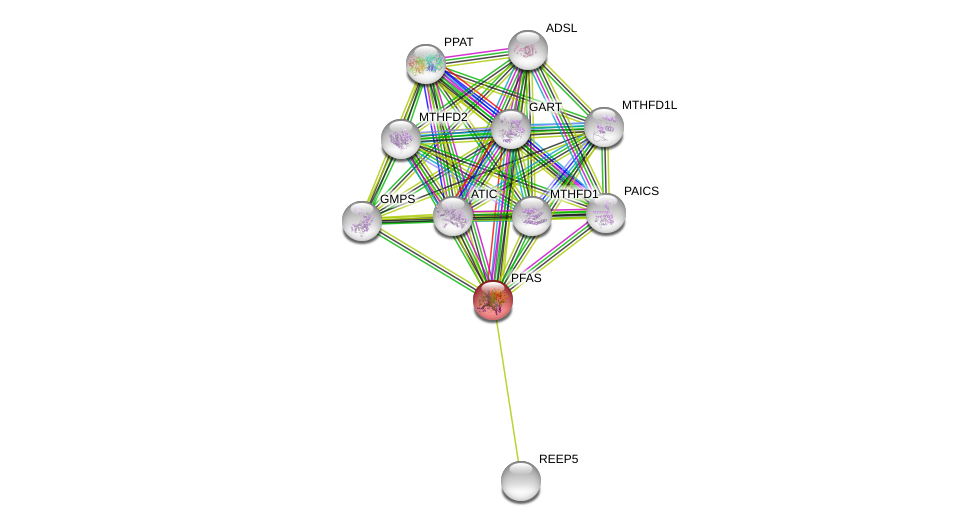
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000178921 | PFAS | 84 | 62.887 | WBGene00008654 | F10F2.2 | 99 | 46.520 | Caenorhabditis_elegans |
| ENSG00000178921 | PFAS | 94 | 35.464 | YGR061C | 93 | 35.464 | Saccharomyces_cerevisiae |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0004642 | phosphoribosylformylglycinamidine synthase activity | 10548741. | IDA | Function |
| GO:0004642 | phosphoribosylformylglycinamidine synthase activity | - | TAS | Function |
| GO:0005524 | ATP binding | - | IEA | Function |
| GO:0005737 | cytoplasm | 21873635. | IBA | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0006189 | 'de novo' IMP biosynthetic process | - | IEA | Process |
| GO:0006189 | 'de novo' IMP biosynthetic process | 10548741. | NAS | Process |
| GO:0006541 | glutamine metabolic process | - | IEA | Process |
| GO:0009168 | purine ribonucleoside monophosphate biosynthetic process | - | TAS | Process |
| GO:0042493 | response to drug | - | IEA | Process |
| GO:0046872 | metal ion binding | - | IEA | Function |
| GO:0070062 | extracellular exosome | 20458337. | HDA | Component |
| GO:0097065 | anterior head development | - | IEA | Process |