
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000327134 | PAK2-201 | 6139 | XM_011512870 | ENSP00000314067 | 524 (aa) | XP_011511172 | Q13177 |
| ENST00000426668 | PAK2-202 | 689 | - | ENSP00000402927 | 221 (aa) | - | H7C1X3 |
| ENST00000481344 | PAK2-203 | 371 | - | - | - (aa) | - | - |
| Pathway ID | Pathway Name | Source |
|---|---|---|
| hsa04010 | MAPK signaling pathway | KEGG |
| hsa04012 | ErbB signaling pathway | KEGG |
| hsa04014 | Ras signaling pathway | KEGG |
| hsa04360 | Axon guidance | KEGG |
| hsa04510 | Focal adhesion | KEGG |
| hsa04660 | T cell receptor signaling pathway | KEGG |
| hsa04810 | Regulation of actin cytoskeleton | KEGG |
| hsa05170 | Human immunodeficiency virus 1 infection | KEGG |
| hsa05211 | Renal cell carcinoma | KEGG |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000180370 | rs7626444 | 3 | 196778031 | C | Monocyte count | 27863252 | [0.016-0.03] unit decrease | 0.02319992 | EFO_0005091 |
| ENSG00000180370 | rs843532 | 3 | 196755340 | ? | Glucose homeostasis traits | 25524916 | [0.055-0.525] unit increase | 0.29 | EFO_0004471|EFO_0006896 |
| ENSG00000180370 | rs2084385 | 3 | 196826996 | ? | Total ventricular volume (Alzheimer's disease interaction) | 21116278 | [NR] unit increase | 0.006 | EFO_0000249|EFO_0006930 |
| ENSG00000180370 | rs10446497 | 3 | 196819658 | G | Schizophrenia | 26198764 | [NR] | 1.0526316 | EFO_0000692 |
| ENSG00000180370 | rs13092376 | 3 | 196789417 | C | Lymphocyte counts | 27863252 | [0.028-0.043] unit decrease | 0.03564678 | EFO_0004587 |
| ENSG00000180370 | rs13434265 | 3 | 196762551 | T | White blood cell count | 27863252 | [0.023-0.038] unit decrease | 0.0303966 | EFO_0004308 |
| ENSG00000180370 | rs11294619 | 3 | 196784238 | T | Plateletcrit | 27863252 | [0.025-0.04] unit decrease | 0.03266217 | EFO_0007985 |
| ENSG00000180370 | rs11294619 | 3 | 196784238 | T | Platelet count | 27863252 | [0.017-0.032] unit decrease | 0.02457575 | EFO_0004309 |
| ENSG00000180370 | rs9872035 | 3 | 196767831 | C | Atrial fibrillation | 29892015 | [1.03-1.06] | 1.04 | EFO_0000275 |
| ENSG00000180370 | rs9829778 | 3 | 196822218 | A | Sum eosinophil basophil counts | 27863252 | [0.016-0.03] unit increase | 0.0230922 | EFO_0005090|EFO_0004842 |
| ENSG00000180370 | rs7630852 | 3 | 196781680 | A | Eosinophil counts | 27863252 | [0.015-0.029] unit decrease | 0.02171766 | EFO_0004842 |
| ENSG00000180370 | rs13092376 | 3 | 196789417 | ? | Red cell distribution width | 30595370 | EFO_0005192 | ||
| ENSG00000180370 | rs2089979 | 3 | 196774542 | ? | White blood cell count | 30595370 | EFO_0004308 | ||
| ENSG00000180370 | rs9829778 | 3 | 196822218 | ? | Eosinophil counts | 30595370 | EFO_0004842 |
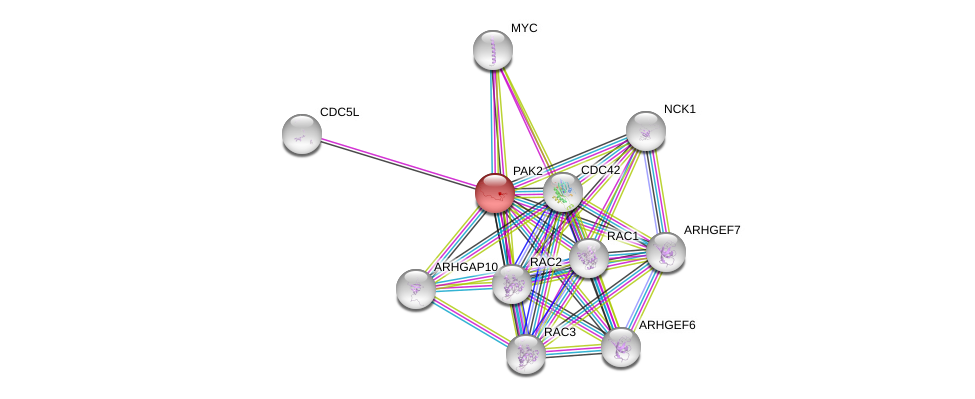
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0002223 | stimulatory C-type lectin receptor signaling pathway | - | TAS | Process |
| GO:0004672 | protein kinase activity | 10748018. | TAS | Function |
| GO:0004674 | protein serine/threonine kinase activity | 11278486. | EXP | Function |
| GO:0004674 | protein serine/threonine kinase activity | 21873635. | IBA | Function |
| GO:0004674 | protein serine/threonine kinase activity | 11121037.11805089. | IDA | Function |
| GO:0004674 | protein serine/threonine kinase activity | - | TAS | Function |
| GO:0005515 | protein binding | 10607567.16374509.16527308.16787945.16837009.18716323.19240112.19264842.19435845.20696164.20936779.21516116.21555521.22705156.25416956. | IPI | Function |
| GO:0005524 | ATP binding | - | IEA | Function |
| GO:0005634 | nucleus | - | IEA | Component |
| GO:0005737 | cytoplasm | 21873635. | IBA | Component |
| GO:0005737 | cytoplasm | 25468996. | IDA | Component |
| GO:0005829 | cytosol | 8625410. | IDA | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0005886 | plasma membrane | - | TAS | Component |
| GO:0006468 | protein phosphorylation | 11805089. | IDA | Process |
| GO:0006469 | negative regulation of protein kinase activity | 10748018. | TAS | Process |
| GO:0006915 | apoptotic process | - | IEA | Process |
| GO:0007165 | signal transduction | 7744004. | TAS | Process |
| GO:0014069 | postsynaptic density | - | IEA | Component |
| GO:0016310 | phosphorylation | 21555521. | IDA | Process |
| GO:0016477 | cell migration | 21873635. | IBA | Process |
| GO:0018105 | peptidyl-serine phosphorylation | 11121037. | IDA | Process |
| GO:0019901 | protein kinase binding | 11121037. | IPI | Function |
| GO:0023014 | signal transduction by protein phosphorylation | 21873635. | IBA | Process |
| GO:0030296 | protein tyrosine kinase activator activity | 11121037. | IDA | Function |
| GO:0031098 | stress-activated protein kinase signaling cascade | 21873635. | IBA | Process |
| GO:0031267 | small GTPase binding | 8625410. | IPI | Function |
| GO:0031295 | T cell costimulation | - | TAS | Process |
| GO:0032147 | activation of protein kinase activity | 21873635. | IBA | Process |
| GO:0035722 | interleukin-12-mediated signaling pathway | - | TAS | Process |
| GO:0038095 | Fc-epsilon receptor signaling pathway | - | TAS | Process |
| GO:0040008 | regulation of growth | - | IEA | Process |
| GO:0042802 | identical protein binding | 16837009. | IPI | Function |
| GO:0043066 | negative regulation of apoptotic process | 21555521. | IMP | Process |
| GO:0043408 | regulation of MAPK cascade | 21873635. | IBA | Process |
| GO:0045296 | cadherin binding | 25468996. | HDA | Function |
| GO:0046777 | protein autophosphorylation | 11805089. | IDA | Process |
| GO:0048010 | vascular endothelial growth factor receptor signaling pathway | - | TAS | Process |
| GO:0048365 | Rac GTPase binding | 21873635. | IBA | Function |
| GO:0048365 | Rac GTPase binding | 8625410. | IPI | Function |
| GO:0048471 | perinuclear region of cytoplasm | - | IEA | Component |
| GO:0050690 | regulation of defense response to virus by virus | - | TAS | Process |
| GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation | 11121037. | IDA | Process |
| GO:0050852 | T cell receptor signaling pathway | - | TAS | Process |
| GO:0060996 | dendritic spine development | - | IEA | Process |
| GO:0061098 | positive regulation of protein tyrosine kinase activity | - | IEA | Process |
| GO:0071407 | cellular response to organic cyclic compound | - | IEA | Process |
| GO:0098978 | glutamatergic synapse | - | IEA | Component |
| GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway | 19240112. | IMP | Process |
| GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis | 21873635. | IBA | Process |
| GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis | 21555521. | IDA | Process |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| DLBC | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRP | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| BLCA | |||
| GBM | |||
| KIRC | |||
| KIRP | |||
| LGG | |||
| LUAD | |||
| PAAD | |||
| PRAD | |||
| SARC |
| Cancer | P-value | Q-value |
|---|---|---|
| THYM | ||
| KIRC | ||
| MESO | ||
| ACC | ||
| SKCM | ||
| PRAD | ||
| KIRP | ||
| PAAD | ||
| KICH | ||
| UCEC | ||
| LIHC | ||
| LGG | ||
| LUAD |