
| PID | Title | Article | Time | Author | Doi |
|---|---|---|---|---|---|
| 23938467 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000595811 | RUVBL2-207 | 572 | - | ENSP00000469760 | 44 (aa) | - | M0QYD8 |
| ENST00000639391 | RUVBL2-213 | 120 | - | ENSP00000492564 | 39 (aa) | - | A0A1W2PS22 |
| ENST00000595002 | RUVBL2-205 | 546 | - | - | - (aa) | - | - |
| ENST00000596837 | RUVBL2-209 | 682 | - | ENSP00000469189 | 109 (aa) | - | M0QXI6 |
| ENST00000593570 | RUVBL2-202 | 813 | - | ENSP00000469488 | 39 (aa) | - | M0QXZ7 |
| ENST00000598768 | RUVBL2-210 | 497 | - | - | - (aa) | - | - |
| ENST00000221413 | RUVBL2-201 | 1478 | - | ENSP00000221413 | 259 (aa) | - | X6R2L4 |
| ENST00000595090 | RUVBL2-206 | 2009 | - | ENSP00000473172 | 463 (aa) | - | Q9Y230 |
| ENST00000601968 | RUVBL2-211 | 1641 | XM_011526330 | ENSP00000471524 | 354 (aa) | XP_011524632 | M0R0Y3 |
| ENST00000596247 | RUVBL2-208 | 1563 | - | ENSP00000471538 | 50 (aa) | - | M0R0Z0 |
| ENST00000640699 | RUVBL2-214 | 153 | - | ENSP00000492610 | 50 (aa) | - | A0A1W2PS48 |
| ENST00000594017 | RUVBL2-203 | 959 | - | - | - (aa) | - | - |
| ENST00000627972 | RUVBL2-212 | 1065 | - | ENSP00000486242 | 354 (aa) | - | M0R0Y3 |
| ENST00000594338 | RUVBL2-204 | 1853 | - | - | - (aa) | - | - |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000183207 | Follicle Stimulating Hormone | 4E-8 | 24049095 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000183207 | rs75287599 | 19 | 49013883 | T | Blood protein levels | 29875488 | [0.33-0.53] unit decrease | 0.43 | EFO_0007937 |
| ENSG00000183207 | rs79785970 | 19 | 49014074 | G | Blood protein levels | 29875488 | [1.03-1.19] unit increase | 1.11 | EFO_0007937 |
| ENSG00000183207 | rs139643250 | 19 | 49013889 | T | Blood protein levels | 29875488 | [0.58-0.78] unit decrease | 0.68 | EFO_0007937 |
| ENSG00000183207 | rs3764618 | 19 | 48993898 | ? | Follicule stimulating hormone | 24049095 | EFO_0004768 | ||
| ENSG00000183207 | rs753307 | 19 | 49015227 | C | Blood protein levels | 28240269 | [0.19-0.32] unit increase | 0.256 | EFO_0008156 |
| ENSG00000183207 | rs79502742 | 19 | 48997109 | G | Blood protein levels | 30072576 | [NR] unit decrease | 0.9031508 | EFO_0007937 |
| ENSG00000183207 | rs79502742 | 19 | 48997109 | G | Blood protein levels | 30072576 | [NR] unit increase | 0.7314155 | EFO_0007937 |
| ENSG00000183207 | rs79502742 | 19 | 48997109 | G | Blood protein levels | 30072576 | [0.54-0.65] unit increase | 0.594 | EFO_0007937 |
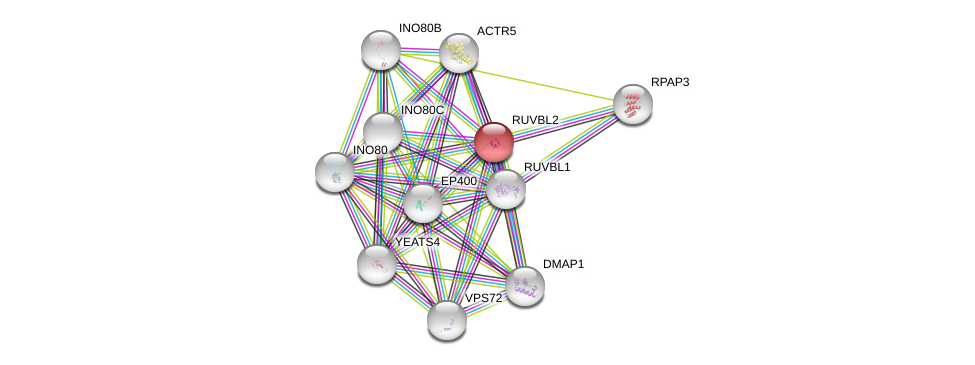
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000183207 | RUVBL2 | 100 | 79.817 | FBgn0040075 | rept | 94 | 78.146 | Drosophila_melanogaster |
| ENSG00000183207 | RUVBL2 | 92 | 57.000 | ENSG00000175792 | RUVBL1 | 100 | 51.908 | Homo_sapiens |
| ENSG00000183207 | RUVBL2 | 100 | 100.000 | ENSMUSG00000003868 | Ruvbl2 | 100 | 99.568 | Mus_musculus |
| ENSG00000183207 | RUVBL2 | 92 | 57.000 | ENSMUSG00000030079 | Ruvbl1 | 100 | 47.761 | Mus_musculus |
| ENSG00000183207 | RUVBL2 | 99 | 78.704 | YPL235W | RVB2 | 94 | 68.764 | Saccharomyces_cerevisiae |
| ENSG00000183207 | RUVBL2 | 92 | 53.000 | YDR190C | RVB1 | 94 | 42.597 | Saccharomyces_cerevisiae |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000492 | box C/D snoRNP assembly | 21873635. | IBA | Process |
| GO:0000812 | Swr1 complex | 21873635. | IBA | Component |
| GO:0000812 | Swr1 complex | 24463511. | IDA | Component |
| GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding | 23637611. | IDA | Function |
| GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding | 23637611. | IDA | Function |
| GO:0001094 | TFIID-class transcription factor complex binding | - | IEA | Function |
| GO:0003678 | DNA helicase activity | 10524211. | IDA | Function |
| GO:0003714 | transcription corepressor activity | 11080158. | IDA | Function |
| GO:0004003 | ATP-dependent DNA helicase activity | 21873635. | IBA | Function |
| GO:0005515 | protein binding | 10966108.11080158.16189514.16230350.17636026.18026119.18087039.18358808.19299493.19433865.20371770.20864032.25416956.27705803.29601588. | IPI | Function |
| GO:0005524 | ATP binding | - | IEA | Function |
| GO:0005634 | nucleus | 10524211.18026119.19433865. | IDA | Component |
| GO:0005654 | nucleoplasm | - | IDA | Component |
| GO:0005654 | nucleoplasm | - | TAS | Component |
| GO:0005719 | nuclear euchromatin | 23637611. | IDA | Component |
| GO:0005737 | cytoplasm | 19433865. | IDA | Component |
| GO:0005813 | centrosome | - | IDA | Component |
| GO:0005829 | cytosol | - | IDA | Component |
| GO:0006281 | DNA repair | - | IEA | Process |
| GO:0006310 | DNA recombination | - | IEA | Process |
| GO:0006338 | chromatin remodeling | 21873635. | IBA | Process |
| GO:0006338 | chromatin remodeling | 23637611. | IMP | Process |
| GO:0006357 | regulation of transcription by RNA polymerase II | 21873635. | IBA | Process |
| GO:0006457 | protein folding | 10524211. | TAS | Process |
| GO:0008013 | beta-catenin binding | 11080158. | IDA | Function |
| GO:0016020 | membrane | - | IEA | Component |
| GO:0016363 | nuclear matrix | - | IEA | Component |
| GO:0016573 | histone acetylation | 21873635. | IBA | Process |
| GO:0016887 | ATPase activity | 10966108. | IDA | Function |
| GO:0017025 | TBP-class protein binding | 11080158. | IDA | Function |
| GO:0031011 | Ino80 complex | 21873635. | IBA | Component |
| GO:0031011 | Ino80 complex | 18026119.21303910. | IDA | Component |
| GO:0031490 | chromatin DNA binding | 23637611. | IDA | Function |
| GO:0032508 | DNA duplex unwinding | - | IEA | Process |
| GO:0034644 | cellular response to UV | 18026119. | IMP | Process |
| GO:0035066 | positive regulation of histone acetylation | 23637611. | IMP | Process |
| GO:0035267 | NuA4 histone acetyltransferase complex | 21873635. | IBA | Component |
| GO:0035267 | NuA4 histone acetyltransferase complex | 10966108.14966270. | IDA | Component |
| GO:0040008 | regulation of growth | - | IEA | Process |
| GO:0042802 | identical protein binding | 10966108. | IDA | Function |
| GO:0042802 | identical protein binding | 10966108.21988832.22748767. | IPI | Function |
| GO:0042803 | protein homodimerization activity | - | IEA | Function |
| GO:0043141 | ATP-dependent 5'-3' DNA helicase activity | - | IEA | Function |
| GO:0043531 | ADP binding | - | IEA | Function |
| GO:0043967 | histone H4 acetylation | 14966270. | IDA | Process |
| GO:0043968 | histone H2A acetylation | 14966270. | IDA | Process |
| GO:0045944 | positive regulation of transcription by RNA polymerase II | 23637611. | IMP | Process |
| GO:0051082 | unfolded protein binding | 10524211. | TAS | Function |
| GO:0051117 | ATPase binding | - | IEA | Function |
| GO:0070062 | extracellular exosome | 20458337. | HDA | Component |
| GO:0071169 | establishment of protein localization to chromatin | 23637611. | IMP | Process |
| GO:0071339 | MLL1 complex | 15960975. | IDA | Component |
| GO:0071392 | cellular response to estradiol stimulus | 23637611. | IMP | Process |
| GO:0071733 | transcriptional activation by promoter-enhancer looping | 23637611. | IMP | Process |
| GO:0071899 | negative regulation of estrogen receptor binding | 23637611. | IMP | Process |
| GO:0090090 | negative regulation of canonical Wnt signaling pathway | 11080158. | IDA | Process |
| GO:0097255 | R2TP complex | 21873635. | IBA | Component |
| GO:0097255 | R2TP complex | 26711270. | IDA | Component |
| GO:1903507 | negative regulation of nucleic acid-templated transcription | - | IEA | Process |
| GO:1904874 | positive regulation of telomerase RNA localization to Cajal body | 25467444. | HMP | Process |
| GO:1990904 | ribonucleoprotein complex | - | IEA | Component |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| ACC | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CHOL | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| KIRP | |||||||
| LAML | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SARC | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| CESC | |||
| KIRP | |||
| LUAD | |||
| LUSC | |||
| PAAD | |||
| READ | |||
| TGCT | |||
| UCS |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| SARC | ||
| MESO | ||
| ACC | ||
| HNSC | ||
| SKCM | ||
| TGCT | ||
| CESC | ||
| LIHC | ||
| DLBC | ||
| LGG | ||
| LUAD | ||
| UVM |