
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000630307 | SETD3-209 | 1281 | - | ENSP00000486445 | 81 (aa) | - | C9K0W5 |
| ENST00000446066 | SETD3-205 | 1754 | - | ENSP00000396870 | 81 (aa) | - | C9K0W5 |
| ENST00000489770 | SETD3-208 | 855 | - | - | - (aa) | - | - |
| ENST00000331768 | SETD3-202 | 2878 | XM_005268127 | ENSP00000327436 | 594 (aa) | XP_005268184 | Q86TU7 |
| ENST00000357563 | SETD3-203 | 2191 | - | ENSP00000350174 | 81 (aa) | - | C9K0W5 |
| ENST00000453764 | SETD3-206 | 973 | - | ENSP00000398966 | 81 (aa) | - | C9K0W5 |
| ENST00000453938 | SETD3-207 | 545 | - | - | - (aa) | - | - |
| ENST00000329331 | SETD3-201 | 1330 | XM_011537235 | ENSP00000327910 | 296 (aa) | XP_011535537 | Q86TU7 |
| ENST00000436070 | SETD3-204 | 2592 | - | ENSP00000408602 | 292 (aa) | - | Q6NXR6 |
| Pathway ID | Pathway Name | Source |
|---|---|---|
| hsa00310 | Lysine degradation | KEGG |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000183576 | rs148151659 | 14 | 99410185 | C | Waist-to-hip ratio adjusted for BMI (additive genetic model) | 30778226 | [0.35-0.91] unit increase | 0.6329 | EFO_0007788 |
| ENSG00000183576 | rs148151659 | 14 | 99410185 | C | Waist-to-hip ratio adjusted for BMI (additive genetic model) | 30778226 | [0.37-0.95] unit increase | 0.6594 | EFO_0007788 |
| ENSG00000183576 | rs148151659 | 14 | 99410185 | C | Waist-to-hip ratio adjusted for BMI (additive genetic model) | 30778226 | [0.61-1.35] unit increase | 0.9768 | EFO_0007788 |
| ENSG00000183576 | rs148151659 | 14 | 99410185 | C | Waist-to-hip ratio adjusted for BMI (additive genetic model) | 30778226 | [0.58-1.28] unit increase | 0.9283 | EFO_0007788 |
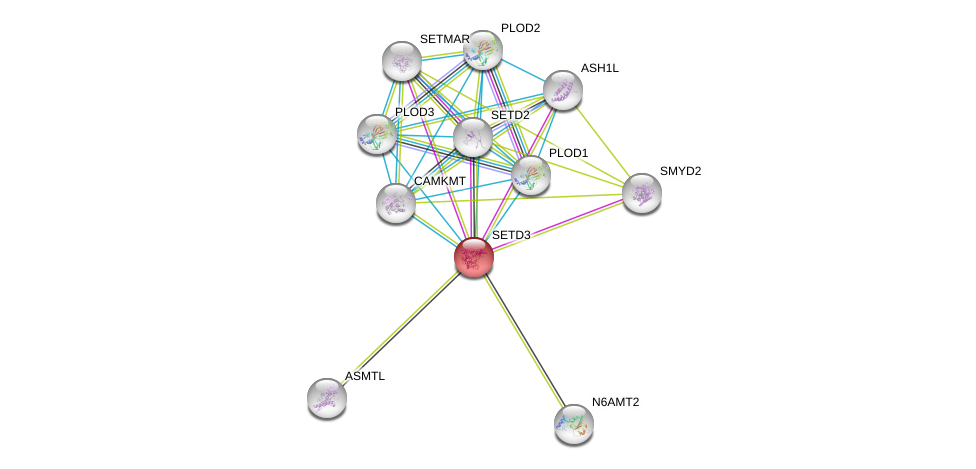
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000790 | nuclear chromatin | 21873635. | IBA | Component |
| GO:0001102 | RNA polymerase II activating transcription factor binding | - | IEA | Function |
| GO:0003713 | transcription coactivator activity | 21873635. | IBA | Function |
| GO:0003713 | transcription coactivator activity | - | ISS | Function |
| GO:0003779 | actin binding | - | IEA | Function |
| GO:0005654 | nucleoplasm | - | TAS | Component |
| GO:0005737 | cytoplasm | 30626964. | IDA | Component |
| GO:0010452 | histone H3-K36 methylation | - | ISS | Process |
| GO:0016279 | protein-lysine N-methyltransferase activity | 21873635. | IBA | Function |
| GO:0018021 | peptidyl-histidine methylation | 30526847.30626964. | IDA | Process |
| GO:0018023 | peptidyl-lysine trimethylation | 21873635. | IBA | Process |
| GO:0018023 | peptidyl-lysine trimethylation | - | ISS | Process |
| GO:0018026 | peptidyl-lysine monomethylation | 21873635. | IBA | Process |
| GO:0018026 | peptidyl-lysine monomethylation | - | ISS | Process |
| GO:0018027 | peptidyl-lysine dimethylation | 21873635. | IBA | Process |
| GO:0018027 | peptidyl-lysine dimethylation | - | ISS | Process |
| GO:0018064 | protein-histidine N-methyltransferase activity | 30526847.30626964. | IDA | Function |
| GO:0030047 | actin modification | 30526847.30626964. | IDA | Process |
| GO:0042800 | histone methyltransferase activity (H3-K4 specific) | 21873635. | IBA | Function |
| GO:0045893 | positive regulation of transcription, DNA-templated | - | ISS | Process |
| GO:0045944 | positive regulation of transcription by RNA polymerase II | 21873635. | IBA | Process |
| GO:0046975 | histone methyltransferase activity (H3-K36 specific) | 21873635. | IBA | Function |
| GO:0046975 | histone methyltransferase activity (H3-K36 specific) | - | ISS | Function |
| GO:0051149 | positive regulation of muscle cell differentiation | - | IEA | Process |
| GO:0051568 | histone H3-K4 methylation | - | IEA | Process |
| GO:0070472 | regulation of uterine smooth muscle contraction | - | ISS | Process |