
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000329858 | Met_10 | PF02475.16 | 1.5e-29 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000521443 | TRMT12-202 | 562 | - | - | - (aa) | - | - |
| ENST00000328599 | TRMT12-201 | 2207 | - | ENSP00000329858 | 448 (aa) | - | Q53H54 |
| ENST00000522518 | TRMT12-203 | 822 | - | ENSP00000429771 | 150 (aa) | - | E5RHH6 |
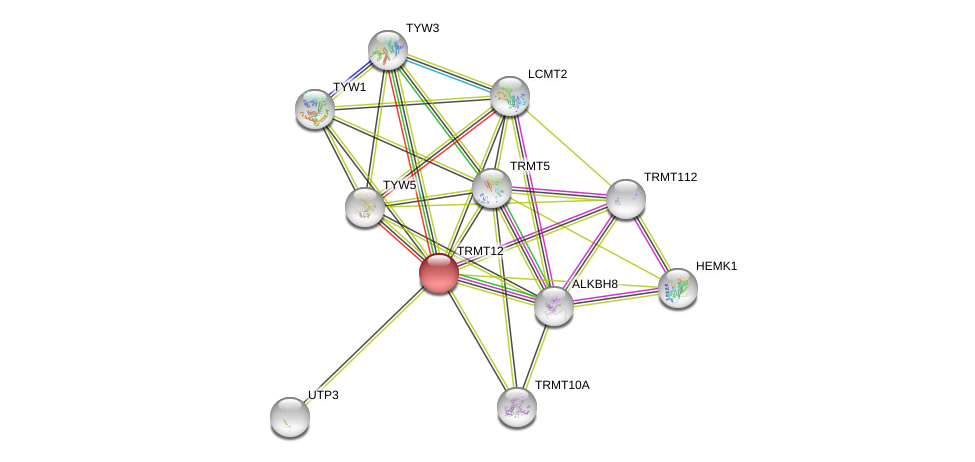
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000183665 | TRMT12 | 92 | 51.090 | ENSAPOG00000011979 | trmt12 | 96 | 51.090 | Acanthochromis_polyacanthus |
| ENSG00000183665 | TRMT12 | 98 | 87.755 | ENSAMEG00000019612 | TRMT12 | 98 | 87.755 | Ailuropoda_melanoleuca |
| ENSG00000183665 | TRMT12 | 74 | 68.750 | ENSACIG00000012982 | trmt12 | 99 | 49.631 | Amphilophus_citrinellus |
| ENSG00000183665 | TRMT12 | 91 | 50.860 | ENSAOCG00000020570 | trmt12 | 96 | 52.163 | Amphiprion_ocellaris |
| ENSG00000183665 | TRMT12 | 91 | 50.860 | ENSAPEG00000006335 | trmt12 | 96 | 52.163 | Amphiprion_percula |
| ENSG00000183665 | TRMT12 | 92 | 50.966 | ENSATEG00000018316 | trmt12 | 97 | 50.966 | Anabas_testudineus |
| ENSG00000183665 | TRMT12 | 92 | 50.121 | ENSACLG00000001008 | trmt12 | 96 | 50.121 | Astatotilapia_calliptera |
| ENSG00000183665 | TRMT12 | 93 | 51.651 | ENSAMXG00000040366 | trmt12 | 97 | 51.651 | Astyanax_mexicanus |
| ENSG00000183665 | TRMT12 | 99 | 83.669 | ENSBTAG00000020398 | TRMT12 | 97 | 83.669 | Bos_taurus |
| ENSG00000183665 | TRMT12 | 100 | 95.238 | ENSCJAG00000022174 | TRMT12 | 100 | 93.527 | Callithrix_jacchus |
| ENSG00000183665 | TRMT12 | 97 | 87.586 | ENSCAFG00000001040 | TRMT12 | 99 | 87.586 | Canis_familiaris |
| ENSG00000183665 | TRMT12 | 97 | 87.586 | ENSCAFG00020006500 | TRMT12 | 99 | 87.586 | Canis_lupus_dingo |
| ENSG00000183665 | TRMT12 | 97 | 85.714 | ENSCHIG00000016574 | TRMT12 | 99 | 85.714 | Capra_hircus |
| ENSG00000183665 | TRMT12 | 98 | 88.095 | ENSCPOG00000026520 | TRMT12 | 100 | 82.472 | Cavia_porcellus |
| ENSG00000183665 | TRMT12 | 100 | 96.205 | ENSCSAG00000018615 | TRMT12 | 100 | 96.205 | Chlorocebus_sabaeus |
| ENSG00000183665 | TRMT12 | 86 | 75.000 | ENSCPBG00000014290 | TRMT12 | 95 | 63.864 | Chrysemys_picta_bellii |
| ENSG00000183665 | TRMT12 | 69 | 45.253 | ENSCING00000019633 | - | 97 | 45.253 | Ciona_intestinalis |
| ENSG00000183665 | TRMT12 | 71 | 47.003 | ENSCSAVG00000000415 | - | 92 | 47.003 | Ciona_savignyi |
| ENSG00000183665 | TRMT12 | 91 | 72.973 | ENSCGRG00001014834 | Trmt12 | 99 | 62.500 | Cricetulus_griseus_chok1gshd |
| ENSG00000183665 | TRMT12 | 92 | 49.395 | ENSCSEG00000002675 | trmt12 | 92 | 49.395 | Cynoglossus_semilaevis |
| ENSG00000183665 | TRMT12 | 93 | 51.190 | ENSCVAG00000005590 | trmt12 | 98 | 51.069 | Cyprinodon_variegatus |
| ENSG00000183665 | TRMT12 | 92 | 49.395 | ENSDARG00000060338 | trmt12 | 95 | 52.593 | Danio_rerio |
| ENSG00000183665 | TRMT12 | 99 | 80.952 | ENSETEG00000009373 | TRMT12 | 99 | 76.957 | Echinops_telfairi |
| ENSG00000183665 | TRMT12 | 98 | 87.699 | ENSEASG00005022441 | TRMT12 | 100 | 87.699 | Equus_asinus_asinus |
| ENSG00000183665 | TRMT12 | 99 | 80.090 | ENSEEUG00000004844 | TRMT12 | 99 | 80.090 | Erinaceus_europaeus |
| ENSG00000183665 | TRMT12 | 92 | 51.202 | ENSELUG00000004240 | trmt12 | 99 | 51.889 | Esox_lucius |
| ENSG00000183665 | TRMT12 | 64 | 62.369 | ENSFALG00000003825 | TRMT12 | 94 | 70.388 | Ficedula_albicollis |
| ENSG00000183665 | TRMT12 | 98 | 85.194 | ENSFDAG00000016647 | TRMT12 | 100 | 85.194 | Fukomys_damarensis |
| ENSG00000183665 | TRMT12 | 91 | 43.521 | ENSFHEG00000017338 | trmt12 | 98 | 43.521 | Fundulus_heteroclitus |
| ENSG00000183665 | TRMT12 | 81 | 61.957 | ENSGALG00000006773 | TRMT12 | 78 | 61.957 | Gallus_gallus |
| ENSG00000183665 | TRMT12 | 79 | 65.625 | ENSGAFG00000020056 | trmt12 | 96 | 49.637 | Gambusia_affinis |
| ENSG00000183665 | TRMT12 | 92 | 49.392 | ENSGACG00000017351 | trmt12 | 100 | 49.392 | Gasterosteus_aculeatus |
| ENSG00000183665 | TRMT12 | 91 | 72.500 | ENSGAGG00000006032 | TRMT12 | 99 | 63.739 | Gopherus_agassizii |
| ENSG00000183665 | TRMT12 | 92 | 50.121 | ENSHBUG00000016412 | trmt12 | 96 | 50.121 | Haplochromis_burtoni |
| ENSG00000183665 | TRMT12 | 97 | 85.550 | ENSHGLG00000020498 | TRMT12 | 99 | 85.550 | Heterocephalus_glaber_female |
| ENSG00000183665 | TRMT12 | 97 | 85.550 | ENSHGLG00100020132 | TRMT12 | 99 | 85.550 | Heterocephalus_glaber_male |
| ENSG00000183665 | TRMT12 | 92 | 48.668 | ENSHCOG00000005050 | trmt12 | 94 | 52.149 | Hippocampus_comes |
| ENSG00000183665 | TRMT12 | 93 | 51.442 | ENSIPUG00000018011 | trmt12 | 89 | 51.442 | Ictalurus_punctatus |
| ENSG00000183665 | TRMT12 | 97 | 86.636 | ENSSTOG00000030617 | TRMT12 | 98 | 86.636 | Ictidomys_tridecemlineatus |
| ENSG00000183665 | TRMT12 | 92 | 49.519 | ENSKMAG00000000891 | trmt12 | 96 | 49.519 | Kryptolebias_marmoratus |
| ENSG00000183665 | TRMT12 | 53 | 55.187 | ENSLBEG00000017456 | - | 88 | 55.187 | Labrus_bergylta |
| ENSG00000183665 | TRMT12 | 92 | 47.952 | ENSLBEG00000028712 | trmt12 | 96 | 47.952 | Labrus_bergylta |
| ENSG00000183665 | TRMT12 | 95 | 67.568 | ENSLACG00000015869 | TRMT12 | 99 | 56.404 | Latimeria_chalumnae |
| ENSG00000183665 | TRMT12 | 85 | 67.568 | ENSLOCG00000015290 | trmt12 | 99 | 54.292 | Lepisosteus_oculatus |
| ENSG00000183665 | TRMT12 | 98 | 86.333 | ENSLAFG00000012519 | TRMT12 | 100 | 86.333 | Loxodonta_africana |
| ENSG00000183665 | TRMT12 | 99 | 96.854 | ENSMMUG00000015490 | TRMT12 | 100 | 96.854 | Macaca_mulatta |
| ENSG00000183665 | TRMT12 | 92 | 50.962 | ENSMAMG00000020311 | trmt12 | 97 | 50.962 | Mastacembelus_armatus |
| ENSG00000183665 | TRMT12 | 92 | 49.637 | ENSMZEG00005007349 | trmt12 | 96 | 49.637 | Maylandia_zebra |
| ENSG00000183665 | TRMT12 | 95 | 58.933 | ENSMGAG00000004174 | TRMT12 | 95 | 58.933 | Meleagris_gallopavo |
| ENSG00000183665 | TRMT12 | 91 | 50.123 | ENSMMOG00000000412 | trmt12 | 95 | 50.123 | Mola_mola |
| ENSG00000183665 | TRMT12 | 99 | 66.516 | ENSMODG00000021403 | - | 99 | 66.516 | Monodelphis_domestica |
| ENSG00000183665 | TRMT12 | 95 | 68.632 | ENSMODG00000011798 | - | 87 | 68.632 | Monodelphis_domestica |
| ENSG00000183665 | TRMT12 | 78 | 67.568 | ENSMALG00000008668 | trmt12 | 96 | 51.090 | Monopterus_albus |
| ENSG00000183665 | TRMT12 | 98 | 80.866 | MGP_CAROLIEiJ_G0019820 | Trmt12 | 98 | 80.866 | Mus_caroli |
| ENSG00000183665 | TRMT12 | 98 | 81.007 | ENSMUSG00000037085 | Trmt12 | 98 | 81.007 | Mus_musculus |
| ENSG00000183665 | TRMT12 | 98 | 80.594 | MGP_PahariEiJ_G0019829 | Trmt12 | 98 | 80.594 | Mus_pahari |
| ENSG00000183665 | TRMT12 | 98 | 80.778 | MGP_SPRETEiJ_G0020717 | Trmt12 | 98 | 80.778 | Mus_spretus |
| ENSG00000183665 | TRMT12 | 98 | 87.528 | ENSMPUG00000019150 | TRMT12 | 99 | 87.528 | Mustela_putorius_furo |
| ENSG00000183665 | TRMT12 | 94 | 90.476 | ENSMLUG00000003753 | TRMT12 | 100 | 84.282 | Myotis_lucifugus |
| ENSG00000183665 | TRMT12 | 98 | 81.321 | ENSNGAG00000014548 | Trmt12 | 100 | 81.321 | Nannospalax_galili |
| ENSG00000183665 | TRMT12 | 92 | 50.365 | ENSNBRG00000021510 | trmt12 | 96 | 50.365 | Neolamprologus_brichardi |
| ENSG00000183665 | TRMT12 | 98 | 72.210 | ENSOPRG00000008067 | TRMT12 | 98 | 72.210 | Ochotona_princeps |
| ENSG00000183665 | TRMT12 | 94 | 88.095 | ENSODEG00000018933 | TRMT12 | 100 | 82.460 | Octodon_degus |
| ENSG00000183665 | TRMT12 | 92 | 50.605 | ENSONIG00000002614 | trmt12 | 96 | 50.605 | Oreochromis_niloticus |
| ENSG00000183665 | TRMT12 | 91 | 78.571 | ENSOANG00000009987 | TRMT12 | 100 | 64.516 | Ornithorhynchus_anatinus |
| ENSG00000183665 | TRMT12 | 97 | 78.440 | ENSOCUG00000026379 | TRMT12 | 97 | 78.440 | Oryctolagus_cuniculus |
| ENSG00000183665 | TRMT12 | 92 | 50.363 | ENSORLG00000001280 | trmt12 | 96 | 50.847 | Oryzias_latipes |
| ENSG00000183665 | TRMT12 | 92 | 50.121 | ENSORLG00020000708 | trmt12 | 96 | 50.605 | Oryzias_latipes_hni |
| ENSG00000183665 | TRMT12 | 92 | 50.605 | ENSORLG00015012344 | trmt12 | 96 | 50.605 | Oryzias_latipes_hsok |
| ENSG00000183665 | TRMT12 | 93 | 50.120 | ENSOMEG00000015267 | trmt12 | 96 | 50.120 | Oryzias_melastigma |
| ENSG00000183665 | TRMT12 | 98 | 84.966 | ENSOGAG00000015845 | TRMT12 | 100 | 84.966 | Otolemur_garnettii |
| ENSG00000183665 | TRMT12 | 93 | 53.444 | ENSPKIG00000004640 | trmt12 | 99 | 53.222 | Paramormyrops_kingsleyae |
| ENSG00000183665 | TRMT12 | 58 | 67.910 | ENSPSIG00000003476 | TRMT12 | 99 | 67.910 | Pelodiscus_sinensis |
| ENSG00000183665 | TRMT12 | 93 | 46.506 | ENSPMGG00000016356 | trmt12 | 94 | 46.506 | Periophthalmus_magnuspinnatus |
| ENSG00000183665 | TRMT12 | 98 | 67.574 | ENSPCIG00000002967 | TRMT12 | 91 | 67.574 | Phascolarctos_cinereus |
| ENSG00000183665 | TRMT12 | 97 | 48.037 | ENSPFOG00000016931 | trmt12 | 99 | 48.037 | Poecilia_formosa |
| ENSG00000183665 | TRMT12 | 92 | 48.681 | ENSPLAG00000020621 | trmt12 | 96 | 48.681 | Poecilia_latipinna |
| ENSG00000183665 | TRMT12 | 92 | 50.121 | ENSPMEG00000000818 | trmt12 | 96 | 50.121 | Poecilia_mexicana |
| ENSG00000183665 | TRMT12 | 79 | 62.500 | ENSPREG00000011621 | trmt12 | 96 | 49.153 | Poecilia_reticulata |
| ENSG00000183665 | TRMT12 | 100 | 98.438 | ENSPPYG00000018866 | TRMT12 | 100 | 98.438 | Pongo_abelii |
| ENSG00000183665 | TRMT12 | 99 | 82.579 | ENSPCAG00000000556 | TRMT12 | 99 | 82.579 | Procavia_capensis |
| ENSG00000183665 | TRMT12 | 100 | 84.598 | ENSPVAG00000009149 | TRMT12 | 100 | 84.598 | Pteropus_vampyrus |
| ENSG00000183665 | TRMT12 | 91 | 49.877 | ENSPNYG00000010696 | trmt12 | 99 | 49.877 | Pundamilia_nyererei |
| ENSG00000183665 | TRMT12 | 92 | 49.763 | ENSPNAG00000019028 | trmt12 | 96 | 49.763 | Pygocentrus_nattereri |
| ENSG00000183665 | TRMT12 | 98 | 82.232 | ENSRNOG00000008978 | Trmt12 | 100 | 82.232 | Rattus_norvegicus |
| ENSG00000183665 | TRMT12 | 97 | 66.975 | ENSSHAG00000003345 | TRMT12 | 96 | 66.975 | Sarcophilus_harrisii |
| ENSG00000183665 | TRMT12 | 95 | 50.698 | ENSSFOG00015006090 | trmt12 | 99 | 50.698 | Scleropages_formosus |
| ENSG00000183665 | TRMT12 | 92 | 50.605 | ENSSMAG00000013247 | trmt12 | 96 | 50.605 | Scophthalmus_maximus |
| ENSG00000183665 | TRMT12 | 91 | 52.580 | ENSSDUG00000006168 | trmt12 | 96 | 52.580 | Seriola_dumerili |
| ENSG00000183665 | TRMT12 | 92 | 52.058 | ENSSLDG00000025007 | trmt12 | 96 | 52.058 | Seriola_lalandi_dorsalis |
| ENSG00000183665 | TRMT12 | 95 | 60.920 | ENSSPUG00000005321 | TRMT12 | 96 | 60.920 | Sphenodon_punctatus |
| ENSG00000183665 | TRMT12 | 92 | 50.121 | ENSSPAG00000009435 | trmt12 | 93 | 50.121 | Stegastes_partitus |
| ENSG00000183665 | TRMT12 | 95 | 55.399 | ENSTGUG00000002280 | TRMT12 | 99 | 55.220 | Taeniopygia_guttata |
| ENSG00000183665 | TRMT12 | 94 | 49.644 | ENSTRUG00000011548 | trmt12 | 95 | 50.000 | Takifugu_rubripes |
| ENSG00000183665 | TRMT12 | 86 | 53.247 | ENSTNIG00000016866 | trmt12 | 88 | 53.247 | Tetraodon_nigroviridis |
| ENSG00000183665 | TRMT12 | 97 | 86.636 | ENSUAMG00000002067 | TRMT12 | 99 | 86.636 | Ursus_americanus |
| ENSG00000183665 | TRMT12 | 99 | 86.682 | ENSVPAG00000006519 | TRMT12 | 99 | 86.682 | Vicugna_pacos |
| ENSG00000183665 | TRMT12 | 96 | 85.880 | ENSVVUG00000016790 | TRMT12 | 97 | 85.880 | Vulpes_vulpes |
| ENSG00000183665 | TRMT12 | 95 | 63.415 | ENSXETG00000009641 | trmt12 | 98 | 54.773 | Xenopus_tropicalis |
| ENSG00000183665 | TRMT12 | 92 | 49.760 | ENSXCOG00000016505 | trmt12 | 99 | 46.824 | Xiphophorus_couchianus |
| ENSG00000183665 | TRMT12 | 92 | 49.517 | ENSXMAG00000025318 | trmt12 | 96 | 49.517 | Xiphophorus_maculatus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0005515 | protein binding | 25416956. | IPI | Function |
| GO:0005737 | cytoplasm | 21873635. | IBA | Component |
| GO:0008175 | tRNA methyltransferase activity | 21873635. | IBA | Function |
| GO:0030488 | tRNA methylation | 21873635. | IBA | Process |
| GO:0031591 | wybutosine biosynthetic process | 21873635. | IBA | Process |
| GO:0102522 | tRNA 4-demethylwyosine alpha-amino-alpha-carboxypropyltransferase activity | - | IEA | Function |