
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
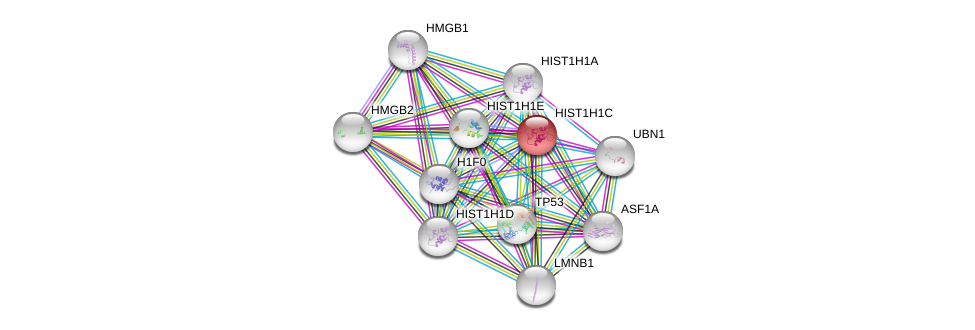
| ENST00000343677 | HIST1H1C-201 | 642 | - | ENSP00000339566 | 213 (aa) | - | P16403 |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000122 | negative regulation of transcription by RNA polymerase II | 21873635. | IBA | Process |
| GO:0000786 | nucleosome | - | IEA | Component |
| GO:0003690 | double-stranded DNA binding | 21873635. | IBA | Function |
| GO:0003723 | RNA binding | 22658674.22681889. | HDA | Function |
| GO:0005515 | protein binding | 21873635. | IBA | Function |
| GO:0005515 | protein binding | 22720776. | IPI | Function |
| GO:0005634 | nucleus | 21873635. | IBA | Component |
| GO:0005634 | nucleus | 21383955.22720776. | IDA | Component |
| GO:0005719 | nuclear euchromatin | 21873635. | IBA | Component |
| GO:0005719 | nuclear euchromatin | 15911621. | IDA | Component |
| GO:0006334 | nucleosome assembly | - | IEA | Process |
| GO:0006355 | regulation of transcription, DNA-templated | 21873635. | IBA | Process |
| GO:0016584 | nucleosome positioning | 21873635. | IBA | Process |
| GO:0030261 | chromosome condensation | 21873635. | IBA | Process |
| GO:0031490 | chromatin DNA binding | 15911621. | IDA | Function |
| GO:0031492 | nucleosomal DNA binding | 21873635. | IBA | Function |
| GO:0031936 | negative regulation of chromatin silencing | 21873635. | IBA | Process |
| GO:0035327 | transcriptionally active chromatin | 21873635. | IBA | Component |
| GO:0045910 | negative regulation of DNA recombination | 21873635. | IBA | Process |
| GO:0080182 | histone H3-K4 trimethylation | 21873635. | IBA | Process |
| GO:0098532 | histone H3-K27 trimethylation | 21873635. | IBA | Process |
| PID | Title | Article | Time | Author | Doi |
|---|---|---|---|---|---|
| 26209608 |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| DLBC | |||||||
| DLBC | |||||||
| DLBC | |||||||
| DLBC | |||||||
| DLBC | |||||||
| DLBC | |||||||
| DLBC | |||||||
| DLBC | |||||||
| DLBC | |||||||
| DLBC | |||||||
| DLBC | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRP | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| MESO | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| READ | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| TGCT | |||||||
| THCA | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| CESC |
| Cancer | P-value | Q-value |
|---|---|---|
| THYM | ||
| MESO | ||
| ACC | ||
| UCS | ||
| PAAD | ||
| BLCA | ||
| CESC | ||
| LAML | ||
| UCEC | ||
| LGG | ||
| UVM | ||
| OV |