
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000344791 | zf-C2H2 | PF00096.26 | 6.3e-80 | 1 | 14 |
| ENSP00000344791 | zf-C2H2 | PF00096.26 | 6.3e-80 | 2 | 14 |
| ENSP00000344791 | zf-C2H2 | PF00096.26 | 6.3e-80 | 3 | 14 |
| ENSP00000344791 | zf-C2H2 | PF00096.26 | 6.3e-80 | 4 | 14 |
| ENSP00000344791 | zf-C2H2 | PF00096.26 | 6.3e-80 | 5 | 14 |
| ENSP00000344791 | zf-C2H2 | PF00096.26 | 6.3e-80 | 6 | 14 |
| ENSP00000344791 | zf-C2H2 | PF00096.26 | 6.3e-80 | 7 | 14 |
| ENSP00000344791 | zf-C2H2 | PF00096.26 | 6.3e-80 | 8 | 14 |
| ENSP00000344791 | zf-C2H2 | PF00096.26 | 6.3e-80 | 9 | 14 |
| ENSP00000344791 | zf-C2H2 | PF00096.26 | 6.3e-80 | 10 | 14 |
| ENSP00000344791 | zf-C2H2 | PF00096.26 | 6.3e-80 | 11 | 14 |
| ENSP00000344791 | zf-C2H2 | PF00096.26 | 6.3e-80 | 12 | 14 |
| ENSP00000344791 | zf-C2H2 | PF00096.26 | 6.3e-80 | 13 | 14 |
| ENSP00000344791 | zf-C2H2 | PF00096.26 | 6.3e-80 | 14 | 14 |
| ENSP00000497540 | zf-C2H2 | PF00096.26 | 4.9e-79 | 1 | 14 |
| ENSP00000497540 | zf-C2H2 | PF00096.26 | 4.9e-79 | 2 | 14 |
| ENSP00000497540 | zf-C2H2 | PF00096.26 | 4.9e-79 | 3 | 14 |
| ENSP00000497540 | zf-C2H2 | PF00096.26 | 4.9e-79 | 4 | 14 |
| ENSP00000497540 | zf-C2H2 | PF00096.26 | 4.9e-79 | 5 | 14 |
| ENSP00000497540 | zf-C2H2 | PF00096.26 | 4.9e-79 | 6 | 14 |
| ENSP00000497540 | zf-C2H2 | PF00096.26 | 4.9e-79 | 7 | 14 |
| ENSP00000497540 | zf-C2H2 | PF00096.26 | 4.9e-79 | 8 | 14 |
| ENSP00000497540 | zf-C2H2 | PF00096.26 | 4.9e-79 | 9 | 14 |
| ENSP00000497540 | zf-C2H2 | PF00096.26 | 4.9e-79 | 10 | 14 |
| ENSP00000497540 | zf-C2H2 | PF00096.26 | 4.9e-79 | 11 | 14 |
| ENSP00000497540 | zf-C2H2 | PF00096.26 | 4.9e-79 | 12 | 14 |
| ENSP00000497540 | zf-C2H2 | PF00096.26 | 4.9e-79 | 13 | 14 |
| ENSP00000497540 | zf-C2H2 | PF00096.26 | 4.9e-79 | 14 | 14 |
| ENSP00000344791 | zf-met | PF12874.7 | 4.4e-11 | 1 | 3 |
| ENSP00000344791 | zf-met | PF12874.7 | 4.4e-11 | 2 | 3 |
| ENSP00000344791 | zf-met | PF12874.7 | 4.4e-11 | 3 | 3 |
| ENSP00000497540 | zf-met | PF12874.7 | 2.8e-10 | 1 | 3 |
| ENSP00000497540 | zf-met | PF12874.7 | 2.8e-10 | 2 | 3 |
| ENSP00000497540 | zf-met | PF12874.7 | 2.8e-10 | 3 | 3 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000647907 | ZNF600-206 | 2608 | - | ENSP00000497989 | 210 (aa) | - | - |
| ENST00000599893 | ZNF600-205 | 600 | - | - | - (aa) | - | - |
| ENST00000594028 | ZNF600-202 | 506 | - | - | - (aa) | - | - |
| ENST00000648893 | ZNF600-207 | 2987 | - | ENSP00000497895 | 204 (aa) | - | - |
| ENST00000648973 | ZNF600-208 | 3645 | XM_017026390 | ENSP00000497540 | 791 (aa) | XP_016881879 | UPI00001D818F |
| ENST00000598369 | ZNF600-204 | 650 | - | - | - (aa) | - | - |
| ENST00000338230 | ZNF600-201 | 3829 | - | ENSP00000344791 | 722 (aa) | - | Q6ZNG1 |
| ENST00000597124 | ZNF600-203 | 333 | - | - | - (aa) | - | - |
| Pathway ID | Pathway Name | Source |
|---|---|---|
| hsa05168 | Herpes simplex virus 1 infection | KEGG |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000189190 | rs10404486 | 19 | 52780882 | ? | Phospholipid levels (plasma) | 22359512 | [NR] % increase | 0.8 | EFO_0004639 |
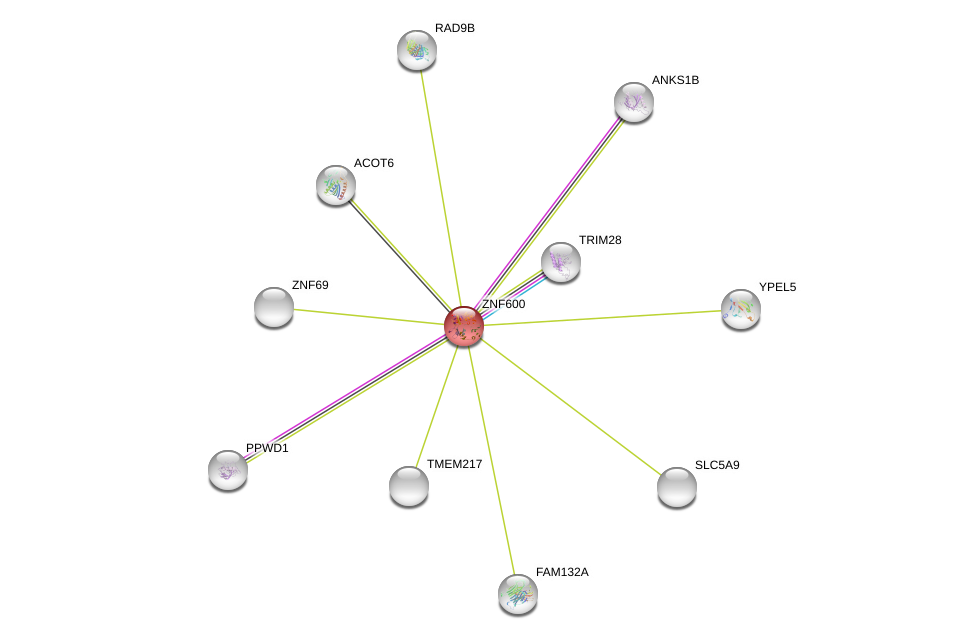
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000189190 | ZNF600 | 99 | 87.216 | ENSCATG00000036192 | - | 99 | 87.216 | Cercocebus_atys |
| ENSG00000189190 | ZNF600 | 100 | 88.089 | ENSCSAG00000019650 | - | 100 | 88.089 | Chlorocebus_sabaeus |
| ENSG00000189190 | ZNF600 | 99 | 86.525 | ENSCANG00000042957 | - | 99 | 86.525 | Colobus_angolensis_palliatus |
| ENSG00000189190 | ZNF600 | 99 | 83.333 | ENSGGOG00000000018 | ZNF611 | 99 | 74.610 | Gorilla_gorilla |
| ENSG00000189190 | ZNF600 | 100 | 89.836 | ENSGGOG00000023190 | ZNF600 | 100 | 86.150 | Gorilla_gorilla |
| ENSG00000189190 | ZNF600 | 100 | 81.431 | ENSMFAG00000030933 | - | 100 | 82.271 | Macaca_fascicularis |
| ENSG00000189190 | ZNF600 | 100 | 87.950 | ENSMFAG00000033366 | - | 100 | 88.258 | Macaca_fascicularis |
| ENSG00000189190 | ZNF600 | 99 | 87.358 | ENSMMUG00000006472 | - | 99 | 87.358 | Macaca_mulatta |
| ENSG00000189190 | ZNF600 | 100 | 88.089 | ENSMMUG00000017044 | - | 100 | 88.092 | Macaca_mulatta |
| ENSG00000189190 | ZNF600 | 99 | 87.421 | ENSMNEG00000045369 | - | 100 | 87.663 | Macaca_nemestrina |
| ENSG00000189190 | ZNF600 | 99 | 86.635 | ENSMLEG00000038583 | - | 96 | 86.635 | Mandrillus_leucophaeus |
| ENSG00000189190 | ZNF600 | 99 | 87.801 | ENSNLEG00000006047 | - | 100 | 87.801 | Nomascus_leucogenys |
| ENSG00000189190 | ZNF600 | 99 | 95.429 | ENSPPAG00000033496 | ZNF600 | 100 | 95.429 | Pan_paniscus |
| ENSG00000189190 | ZNF600 | 100 | 98.199 | ENSPTRG00000011412 | ZNF600 | 100 | 98.199 | Pan_troglodytes |
| ENSG00000189190 | ZNF600 | 100 | 81.431 | ENSPANG00000010431 | - | 100 | 82.271 | Papio_anubis |
| ENSG00000189190 | ZNF600 | 100 | 88.089 | ENSPANG00000020931 | - | 100 | 88.387 | Papio_anubis |
| ENSG00000189190 | ZNF600 | 99 | 86.164 | ENSPPYG00000029892 | - | 100 | 86.792 | Pongo_abelii |
| ENSG00000189190 | ZNF600 | 100 | 87.358 | ENSRROG00000037435 | - | 100 | 87.358 | Rhinopithecus_roxellana |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000981 | DNA-binding transcription factor activity, RNA polymerase II-specific | - | ISA | Function |
| GO:0003677 | DNA binding | - | IEA | Function |
| GO:0005634 | nucleus | - | IEA | Component |
| GO:0006357 | regulation of transcription by RNA polymerase II | - | IEA | Process |
| GO:0046872 | metal ion binding | - | IEA | Function |