
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000384082 | SAP | PF02037.27 | 7.9e-09 | 1 | 1 |
| ENSP00000403679 | SAP | PF02037.27 | 7.9e-09 | 1 | 1 |
| ENSP00000384941 | SAP | PF02037.27 | 9.5e-09 | 1 | 1 |
| ENSP00000352257 | SAP | PF02037.27 | 1.1e-08 | 1 | 1 |
| ENSP00000353192 | SAP | PF02037.27 | 1.1e-08 | 1 | 1 |
| ENSP00000384257 | SAP | PF02037.27 | 1.1e-08 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000405878 | XRCC6-205 | 2200 | - | ENSP00000384257 | 609 (aa) | - | P12956 |
| ENST00000405506 | XRCC6-204 | 1958 | - | ENSP00000384082 | 559 (aa) | - | B1AHC9 |
| ENST00000428575 | XRCC6-206 | 2148 | - | ENSP00000403679 | 559 (aa) | - | B1AHC9 |
| ENST00000464116 | XRCC6-207 | 359 | - | - | - (aa) | - | - |
| ENST00000478914 | XRCC6-208 | 550 | - | - | - (aa) | - | - |
| ENST00000360079 | XRCC6-202 | 2296 | - | ENSP00000353192 | 609 (aa) | - | P12956 |
| ENST00000402580 | XRCC6-203 | 2006 | - | ENSP00000384941 | 568 (aa) | - | P12956 |
| ENST00000359308 | XRCC6-201 | 2709 | - | ENSP00000352257 | 609 (aa) | - | P12956 |
| Pathway ID | Pathway Name | Source |
|---|---|---|
| hsa03450 | Non-homologous end-joining | KEGG |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000196419 | rs28741121 | 22 | 41629819 | G | Highest math class taken | 30038396 | [0.011-0.023] unit decrease | 0.0172 | EFO_0004875 |
| ENSG00000196419 | rs28741121 | 22 | 41629819 | A | Educational attainment (MTAG) | 30038396 | [0.0092-0.0158] unit increase | 0.0125 | EFO_0004784 |
| ENSG00000196419 | rs28741121 | 22 | 41629819 | A | Highest math class taken (MTAG) | 30038396 | [0.014-0.023] unit increase | 0.0182 | EFO_0004875 |
| ENSG00000196419 | rs73161324 | 22 | 41642782 | T | Pulse pressure | 28135244 | [0.36-0.63] unit increase | 0.496 | EFO_0005763 |
| ENSG00000196419 | rs28741121 | 22 | 41629819 | A | Intelligence | 29942086 | z-score increase | 6.609 | EFO_0004337 |
| ENSG00000196419 | rs73161324 | 22 | 41642782 | T | Breast cancer | 29059683 | [1.02-1.09] (Oncoarray) | 1.06 | EFO_0000305 |
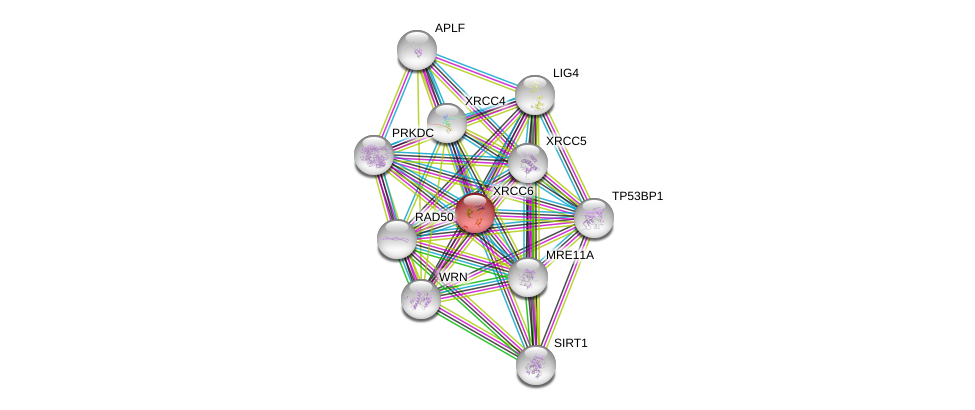
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000196419 | XRCC6 | 99 | 56.414 | ENSAPOG00000012704 | xrcc6 | 99 | 56.414 | Acanthochromis_polyacanthus |
| ENSG00000196419 | XRCC6 | 99 | 56.086 | ENSACIG00000021277 | xrcc6 | 99 | 56.086 | Amphilophus_citrinellus |
| ENSG00000196419 | XRCC6 | 99 | 55.337 | ENSAOCG00000021344 | xrcc6 | 99 | 55.410 | Amphiprion_ocellaris |
| ENSG00000196419 | XRCC6 | 99 | 55.665 | ENSAPEG00000020712 | xrcc6 | 99 | 55.738 | Amphiprion_percula |
| ENSG00000196419 | XRCC6 | 99 | 56.250 | ENSATEG00000007119 | xrcc6 | 99 | 56.086 | Anabas_testudineus |
| ENSG00000196419 | XRCC6 | 97 | 71.402 | ENSAPLG00000004469 | XRCC6 | 99 | 70.962 | Anas_platyrhynchos |
| ENSG00000196419 | XRCC6 | 100 | 85.714 | ENSANAG00000035230 | - | 100 | 85.714 | Aotus_nancymaae |
| ENSG00000196419 | XRCC6 | 100 | 98.686 | ENSANAG00000018684 | XRCC6 | 100 | 98.686 | Aotus_nancymaae |
| ENSG00000196419 | XRCC6 | 100 | 83.713 | ENSANAG00000023445 | - | 100 | 83.713 | Aotus_nancymaae |
| ENSG00000196419 | XRCC6 | 100 | 87.192 | ENSANAG00000020017 | - | 91 | 96.535 | Aotus_nancymaae |
| ENSG00000196419 | XRCC6 | 99 | 56.414 | ENSACLG00000018608 | xrcc6 | 99 | 56.250 | Astatotilapia_calliptera |
| ENSG00000196419 | XRCC6 | 99 | 57.516 | ENSAMXG00000007545 | xrcc6 | 99 | 57.353 | Astyanax_mexicanus |
| ENSG00000196419 | XRCC6 | 100 | 98.522 | ENSCJAG00000045512 | XRCC6 | 100 | 98.522 | Callithrix_jacchus |
| ENSG00000196419 | XRCC6 | 99 | 87.007 | ENSTSYG00000011348 | XRCC6 | 99 | 87.007 | Carlito_syrichta |
| ENSG00000196419 | XRCC6 | 100 | 92.447 | ENSCCAG00000030041 | - | 95 | 92.447 | Cebus_capucinus |
| ENSG00000196419 | XRCC6 | 100 | 79.310 | ENSCCAG00000035711 | - | 100 | 79.310 | Cebus_capucinus |
| ENSG00000196419 | XRCC6 | 90 | 98.438 | ENSCCAG00000030468 | - | 100 | 98.438 | Cebus_capucinus |
| ENSG00000196419 | XRCC6 | 100 | 79.310 | ENSCCAG00000037663 | - | 97 | 78.654 | Cebus_capucinus |
| ENSG00000196419 | XRCC6 | 100 | 99.179 | ENSCATG00000042366 | XRCC6 | 100 | 99.179 | Cercocebus_atys |
| ENSG00000196419 | XRCC6 | 99 | 85.667 | ENSCLAG00000016669 | XRCC6 | 99 | 85.667 | Chinchilla_lanigera |
| ENSG00000196419 | XRCC6 | 100 | 99.179 | ENSCSAG00000005784 | XRCC6 | 100 | 99.179 | Chlorocebus_sabaeus |
| ENSG00000196419 | XRCC6 | 99 | 74.056 | ENSCPBG00000020535 | XRCC6 | 99 | 74.220 | Chrysemys_picta_bellii |
| ENSG00000196419 | XRCC6 | 97 | 35.949 | ENSCING00000000633 | - | 95 | 35.678 | Ciona_intestinalis |
| ENSG00000196419 | XRCC6 | 100 | 99.179 | ENSCANG00000031032 | XRCC6 | 100 | 99.179 | Colobus_angolensis_palliatus |
| ENSG00000196419 | XRCC6 | 99 | 77.138 | ENSCGRG00001019029 | Xrcc6 | 99 | 77.138 | Cricetulus_griseus_chok1gshd |
| ENSG00000196419 | XRCC6 | 99 | 84.868 | ENSCGRG00000001528 | Xrcc6 | 99 | 84.868 | Cricetulus_griseus_crigri |
| ENSG00000196419 | XRCC6 | 99 | 56.373 | ENSCSEG00000020434 | xrcc6 | 99 | 56.209 | Cynoglossus_semilaevis |
| ENSG00000196419 | XRCC6 | 99 | 56.158 | ENSCVAG00000002189 | xrcc6 | 99 | 56.158 | Cyprinodon_variegatus |
| ENSG00000196419 | XRCC6 | 99 | 58.033 | ENSDARG00000071551 | xrcc6 | 99 | 58.361 | Danio_rerio |
| ENSG00000196419 | XRCC6 | 84 | 76.923 | ENSETEG00000015333 | - | 78 | 76.923 | Echinops_telfairi |
| ENSG00000196419 | XRCC6 | 99 | 85.691 | ENSEASG00005001514 | XRCC6 | 99 | 86.020 | Equus_asinus_asinus |
| ENSG00000196419 | XRCC6 | 99 | 85.197 | ENSECAG00000017563 | XRCC6 | 99 | 86.275 | Equus_caballus |
| ENSG00000196419 | XRCC6 | 100 | 54.560 | ENSELUG00000020301 | xrcc6 | 100 | 54.560 | Esox_lucius |
| ENSG00000196419 | XRCC6 | 99 | 70.328 | ENSFALG00000011908 | XRCC6 | 97 | 69.771 | Ficedula_albicollis |
| ENSG00000196419 | XRCC6 | 99 | 57.190 | ENSFHEG00000016833 | xrcc6 | 99 | 57.026 | Fundulus_heteroclitus |
| ENSG00000196419 | XRCC6 | 99 | 48.768 | ENSGMOG00000013524 | xrcc6 | 99 | 48.768 | Gadus_morhua |
| ENSG00000196419 | XRCC6 | 99 | 69.722 | ENSGALG00000011932 | XRCC6 | 99 | 69.558 | Gallus_gallus |
| ENSG00000196419 | XRCC6 | 99 | 55.065 | ENSGAFG00000008130 | xrcc6 | 99 | 55.065 | Gambusia_affinis |
| ENSG00000196419 | XRCC6 | 99 | 54.098 | ENSGACG00000004868 | xrcc6 | 99 | 54.098 | Gasterosteus_aculeatus |
| ENSG00000196419 | XRCC6 | 99 | 73.892 | ENSGAGG00000017518 | XRCC6 | 99 | 74.056 | Gopherus_agassizii |
| ENSG00000196419 | XRCC6 | 100 | 99.672 | ENSGGOG00000015885 | XRCC6 | 100 | 99.672 | Gorilla_gorilla |
| ENSG00000196419 | XRCC6 | 96 | 57.196 | ENSHBUG00000023281 | xrcc6 | 99 | 56.506 | Haplochromis_burtoni |
| ENSG00000196419 | XRCC6 | 99 | 84.020 | ENSHGLG00000018078 | XRCC6 | 99 | 84.020 | Heterocephalus_glaber_female |
| ENSG00000196419 | XRCC6 | 99 | 84.020 | ENSHGLG00100011264 | XRCC6 | 99 | 84.020 | Heterocephalus_glaber_male |
| ENSG00000196419 | XRCC6 | 99 | 53.618 | ENSHCOG00000009821 | xrcc6 | 99 | 53.947 | Hippocampus_comes |
| ENSG00000196419 | XRCC6 | 97 | 57.038 | ENSIPUG00000000767 | xrcc6 | 96 | 57.167 | Ictalurus_punctatus |
| ENSG00000196419 | XRCC6 | 99 | 88.158 | ENSSTOG00000009490 | XRCC6 | 99 | 87.993 | Ictidomys_tridecemlineatus |
| ENSG00000196419 | XRCC6 | 99 | 78.618 | ENSJJAG00000017136 | Xrcc6 | 99 | 79.605 | Jaculus_jaculus |
| ENSG00000196419 | XRCC6 | 99 | 56.158 | ENSKMAG00000002141 | xrcc6 | 99 | 55.993 | Kryptolebias_marmoratus |
| ENSG00000196419 | XRCC6 | 99 | 55.556 | ENSLBEG00000002691 | xrcc6 | 99 | 55.556 | Labrus_bergylta |
| ENSG00000196419 | XRCC6 | 90 | 58.824 | ENSLOCG00000011533 | xrcc6 | 99 | 58.824 | Lepisosteus_oculatus |
| ENSG00000196419 | XRCC6 | 100 | 99.179 | ENSMFAG00000003352 | XRCC6 | 100 | 99.179 | Macaca_fascicularis |
| ENSG00000196419 | XRCC6 | 100 | 99.179 | ENSMMUG00000001856 | XRCC6 | 100 | 99.179 | Macaca_mulatta |
| ENSG00000196419 | XRCC6 | 100 | 99.179 | ENSMNEG00000035076 | XRCC6 | 100 | 99.179 | Macaca_nemestrina |
| ENSG00000196419 | XRCC6 | 100 | 99.179 | ENSMLEG00000019869 | XRCC6 | 100 | 99.179 | Mandrillus_leucophaeus |
| ENSG00000196419 | XRCC6 | 96 | 55.331 | ENSMAMG00000003405 | xrcc6 | 96 | 55.403 | Mastacembelus_armatus |
| ENSG00000196419 | XRCC6 | 99 | 54.918 | ENSMAMG00000003368 | xrcc6 | 99 | 55.082 | Mastacembelus_armatus |
| ENSG00000196419 | XRCC6 | 99 | 56.414 | ENSMZEG00005028080 | xrcc6 | 99 | 56.250 | Maylandia_zebra |
| ENSG00000196419 | XRCC6 | 99 | 69.494 | ENSMGAG00000012146 | XRCC6 | 99 | 69.331 | Meleagris_gallopavo |
| ENSG00000196419 | XRCC6 | 99 | 84.539 | ENSMAUG00000017340 | Xrcc6 | 99 | 84.539 | Mesocricetus_auratus |
| ENSG00000196419 | XRCC6 | 99 | 84.375 | ENSMOCG00000015717 | Xrcc6 | 99 | 84.539 | Microtus_ochrogaster |
| ENSG00000196419 | XRCC6 | 99 | 54.680 | ENSMMOG00000003536 | xrcc6 | 99 | 54.680 | Mola_mola |
| ENSG00000196419 | XRCC6 | 99 | 57.401 | ENSMALG00000013912 | xrcc6 | 94 | 57.237 | Monopterus_albus |
| ENSG00000196419 | XRCC6 | 99 | 77.467 | MGP_CAROLIEiJ_G0020067 | Xrcc6 | 99 | 77.467 | Mus_caroli |
| ENSG00000196419 | XRCC6 | 99 | 83.224 | ENSMUSG00000022471 | Xrcc6 | 99 | 83.224 | Mus_musculus |
| ENSG00000196419 | XRCC6 | 99 | 82.401 | MGP_PahariEiJ_G0020069 | Xrcc6 | 99 | 82.401 | Mus_pahari |
| ENSG00000196419 | XRCC6 | 97 | 83.272 | MGP_SPRETEiJ_G0020967 | Xrcc6 | 99 | 83.821 | Mus_spretus |
| ENSG00000196419 | XRCC6 | 99 | 83.553 | ENSNGAG00000013750 | Xrcc6 | 99 | 83.553 | Nannospalax_galili |
| ENSG00000196419 | XRCC6 | 99 | 56.908 | ENSNBRG00000004125 | xrcc6 | 99 | 56.743 | Neolamprologus_brichardi |
| ENSG00000196419 | XRCC6 | 97 | 82.505 | ENSOPRG00000002386 | XRCC6 | 99 | 82.931 | Ochotona_princeps |
| ENSG00000196419 | XRCC6 | 99 | 82.208 | ENSODEG00000015847 | XRCC6 | 99 | 82.372 | Octodon_degus |
| ENSG00000196419 | XRCC6 | 99 | 56.743 | ENSONIG00000019715 | xrcc6 | 99 | 56.908 | Oreochromis_niloticus |
| ENSG00000196419 | XRCC6 | 74 | 79.612 | ENSOANG00000011328 | XRCC6 | 99 | 79.612 | Ornithorhynchus_anatinus |
| ENSG00000196419 | XRCC6 | 100 | 86.207 | ENSOCUG00000006948 | XRCC6 | 100 | 86.535 | Oryctolagus_cuniculus |
| ENSG00000196419 | XRCC6 | 99 | 55.574 | ENSORLG00020007820 | xrcc6 | 99 | 55.574 | Oryzias_latipes_hni |
| ENSG00000196419 | XRCC6 | 99 | 54.976 | ENSORLG00015006406 | xrcc6 | 99 | 55.246 | Oryzias_latipes_hsok |
| ENSG00000196419 | XRCC6 | 99 | 54.812 | ENSOMEG00000008998 | xrcc6 | 99 | 54.812 | Oryzias_melastigma |
| ENSG00000196419 | XRCC6 | 99 | 87.171 | ENSOGAG00000008821 | XRCC6 | 99 | 87.007 | Otolemur_garnettii |
| ENSG00000196419 | XRCC6 | 94 | 100.000 | ENSPPAG00000037167 | XRCC6 | 100 | 100.000 | Pan_paniscus |
| ENSG00000196419 | XRCC6 | 100 | 100.000 | ENSPTRG00000022548 | XRCC6 | 100 | 100.000 | Pan_troglodytes |
| ENSG00000196419 | XRCC6 | 100 | 100.000 | ENSPTRG00000045824 | XRCC6 | 100 | 100.000 | Pan_troglodytes |
| ENSG00000196419 | XRCC6 | 100 | 99.015 | ENSPANG00000001252 | XRCC6 | 100 | 99.015 | Papio_anubis |
| ENSG00000196419 | XRCC6 | 99 | 60.526 | ENSPKIG00000003974 | xrcc6 | 99 | 60.526 | Paramormyrops_kingsleyae |
| ENSG00000196419 | XRCC6 | 99 | 55.537 | ENSPMGG00000014220 | xrcc6 | 100 | 55.702 | Periophthalmus_magnuspinnatus |
| ENSG00000196419 | XRCC6 | 99 | 83.553 | ENSPEMG00000023446 | Xrcc6 | 99 | 83.553 | Peromyscus_maniculatus_bairdii |
| ENSG00000196419 | XRCC6 | 100 | 79.016 | ENSPCIG00000026409 | XRCC6 | 99 | 78.852 | Phascolarctos_cinereus |
| ENSG00000196419 | XRCC6 | 99 | 55.556 | ENSPFOG00000017173 | xrcc6 | 99 | 55.392 | Poecilia_formosa |
| ENSG00000196419 | XRCC6 | 99 | 55.229 | ENSPLAG00000000562 | xrcc6 | 99 | 55.065 | Poecilia_latipinna |
| ENSG00000196419 | XRCC6 | 99 | 55.719 | ENSPMEG00000018478 | xrcc6 | 99 | 55.556 | Poecilia_mexicana |
| ENSG00000196419 | XRCC6 | 99 | 55.537 | ENSPREG00000020957 | xrcc6 | 93 | 55.375 | Poecilia_reticulata |
| ENSG00000196419 | XRCC6 | 100 | 88.670 | ENSPPYG00000011876 | XRCC6 | 100 | 88.670 | Pongo_abelii |
| ENSG00000196419 | XRCC6 | 99 | 85.033 | ENSPVAG00000001168 | XRCC6 | 99 | 84.704 | Pteropus_vampyrus |
| ENSG00000196419 | XRCC6 | 99 | 56.414 | ENSPNYG00000003815 | xrcc6 | 99 | 53.521 | Pundamilia_nyererei |
| ENSG00000196419 | XRCC6 | 99 | 83.388 | ENSRNOG00000006392 | Xrcc6 | 99 | 83.388 | Rattus_norvegicus |
| ENSG00000196419 | XRCC6 | 100 | 99.120 | ENSRBIG00000039794 | XRCC6 | 100 | 99.120 | Rhinopithecus_bieti |
| ENSG00000196419 | XRCC6 | 100 | 99.179 | ENSRROG00000030297 | XRCC6 | 100 | 99.179 | Rhinopithecus_roxellana |
| ENSG00000196419 | XRCC6 | 73 | 85.504 | ENSSBOG00000024724 | - | 100 | 85.504 | Saimiri_boliviensis_boliviensis |
| ENSG00000196419 | XRCC6 | 99 | 78.161 | ENSSHAG00000010790 | XRCC6 | 99 | 77.833 | Sarcophilus_harrisii |
| ENSG00000196419 | XRCC6 | 99 | 60.820 | ENSSFOG00015014409 | xrcc6 | 99 | 60.656 | Scleropages_formosus |
| ENSG00000196419 | XRCC6 | 99 | 56.743 | ENSSMAG00000006496 | xrcc6 | 99 | 56.743 | Scophthalmus_maximus |
| ENSG00000196419 | XRCC6 | 99 | 56.536 | ENSSDUG00000007542 | xrcc6 | 99 | 56.373 | Seriola_dumerili |
| ENSG00000196419 | XRCC6 | 99 | 55.719 | ENSSLDG00000009974 | xrcc6 | 99 | 55.556 | Seriola_lalandi_dorsalis |
| ENSG00000196419 | XRCC6 | 99 | 73.727 | ENSSPUG00000012086 | XRCC6 | 99 | 73.399 | Sphenodon_punctatus |
| ENSG00000196419 | XRCC6 | 99 | 56.373 | ENSSPAG00000015199 | xrcc6 | 99 | 56.209 | Stegastes_partitus |
| ENSG00000196419 | XRCC6 | 97 | 71.956 | ENSTGUG00000009852 | - | 95 | 72.165 | Taeniopygia_guttata |
| ENSG00000196419 | XRCC6 | 99 | 55.263 | ENSTRUG00000017070 | xrcc6 | 99 | 55.099 | Takifugu_rubripes |
| ENSG00000196419 | XRCC6 | 96 | 57.246 | ENSTNIG00000012947 | xrcc6 | 99 | 56.949 | Tetraodon_nigroviridis |
| ENSG00000196419 | XRCC6 | 97 | 78.309 | ENSTBEG00000011736 | XRCC6 | 99 | 78.485 | Tupaia_belangeri |
| ENSG00000196419 | XRCC6 | 99 | 71.217 | ENSTTRG00000006749 | XRCC6 | 99 | 71.546 | Tursiops_truncatus |
| ENSG00000196419 | XRCC6 | 99 | 55.556 | ENSXMAG00000009691 | xrcc6 | 99 | 55.392 | Xiphophorus_maculatus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000723 | telomere maintenance | 21873635. | IBA | Process |
| GO:0000723 | telomere maintenance | 15824061.18710952.24095731. | TAS | Process |
| GO:0000783 | nuclear telomere cap complex | 15100233. | TAS | Component |
| GO:0000784 | nuclear chromosome, telomeric region | 19135898. | HDA | Component |
| GO:0000784 | nuclear chromosome, telomeric region | 21873635. | IBA | Component |
| GO:0002218 | activation of innate immune response | 28712728. | IDA | Process |
| GO:0003677 | DNA binding | 1537839. | NAS | Function |
| GO:0003684 | damaged DNA binding | - | IEA | Function |
| GO:0003690 | double-stranded DNA binding | 21873635. | IBA | Function |
| GO:0003691 | double-stranded telomeric DNA binding | 10409678. | IDA | Function |
| GO:0003723 | RNA binding | 22658674.22681889. | HDA | Function |
| GO:0004003 | ATP-dependent DNA helicase activity | - | IEA | Function |
| GO:0005515 | protein binding | 8621488.10984620.11328876.12145306.12377759.14734561.15075319.15205477.15383276.15383534.15782130.16169070.17124166.17159921.17220478.17283121.17308091.17334224.19303849.20711232.21044950.21070772.21146485.21575865.21679440.22442688.22451927.22504299.23685356.24095731.24610814.24965446.25574025.25749521.25852083.26175416.26359349.26496610.28330616.28959974.30021884. | IPI | Function |
| GO:0005524 | ATP binding | - | IEA | Function |
| GO:0005576 | extracellular region | - | TAS | Component |
| GO:0005634 | nucleus | 10508516. | TAS | Component |
| GO:0005654 | nucleoplasm | - | IDA | Component |
| GO:0005654 | nucleoplasm | - | TAS | Component |
| GO:0005667 | transcription factor complex | 12145306. | IDA | Component |
| GO:0005730 | nucleolus | 27829214. | IDA | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0006266 | DNA ligation | 9826654. | TAS | Process |
| GO:0006303 | double-strand break repair via nonhomologous end joining | 21873635. | IBA | Process |
| GO:0006303 | double-strand break repair via nonhomologous end joining | 20383123.26359349. | IMP | Process |
| GO:0006303 | double-strand break repair via nonhomologous end joining | 15824061. | TAS | Process |
| GO:0006310 | DNA recombination | - | IEA | Process |
| GO:0007420 | brain development | - | IEA | Process |
| GO:0008022 | protein C-terminus binding | 10783163. | IPI | Function |
| GO:0016020 | membrane | 19946888. | HDA | Component |
| GO:0030332 | cyclin binding | 15159402. | IPI | Function |
| GO:0032481 | positive regulation of type I interferon production | - | TAS | Process |
| GO:0032508 | DNA duplex unwinding | - | IEA | Process |
| GO:0032991 | protein-containing complex | 22504299. | IDA | Component |
| GO:0032993 | protein-DNA complex | 22504299. | IDA | Component |
| GO:0034774 | secretory granule lumen | - | TAS | Component |
| GO:0042162 | telomeric DNA binding | 21873635. | IBA | Function |
| GO:0043312 | neutrophil degranulation | - | TAS | Process |
| GO:0043564 | Ku70:Ku80 complex | 21873635. | IBA | Component |
| GO:0043564 | Ku70:Ku80 complex | 20383123.24095731.26359349. | IDA | Component |
| GO:0044212 | transcription regulatory region DNA binding | 18809223. | IDA | Function |
| GO:0044877 | protein-containing complex binding | 12377759. | IPI | Function |
| GO:0045087 | innate immune response | - | IEA | Process |
| GO:0045860 | positive regulation of protein kinase activity | 22504299. | IDA | Process |
| GO:0045892 | negative regulation of transcription, DNA-templated | 8621488. | IMP | Process |
| GO:0045893 | positive regulation of transcription, DNA-templated | 12145306. | IDA | Process |
| GO:0045893 | positive regulation of transcription, DNA-templated | 18809223. | IMP | Process |
| GO:0045944 | positive regulation of transcription by RNA polymerase II | 18809223. | IMP | Process |
| GO:0048660 | regulation of smooth muscle cell proliferation | 25852083. | IMP | Process |
| GO:0051290 | protein heterotetramerization | 24095731. | IDA | Process |
| GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity | 20383123. | IMP | Function |
| GO:0070419 | nonhomologous end joining complex | 20383123.25941166. | IDA | Component |
| GO:0071475 | cellular hyperosmotic salinity response | - | IEA | Process |
| GO:0071480 | cellular response to gamma radiation | 21873635. | IBA | Process |
| GO:0071480 | cellular response to gamma radiation | 26359349. | IDA | Process |
| GO:0071481 | cellular response to X-ray | 21873635. | IBA | Process |
| GO:0075713 | establishment of integrated proviral latency | - | TAS | Process |
| GO:0097680 | double-strand break repair via classical nonhomologous end joining | 24095731. | IDA | Process |
| GO:1904813 | ficolin-1-rich granule lumen | - | TAS | Component |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRP | |||||||
| KIRP | |||||||
| LAML | |||||||
| LAML | |||||||
| LAML | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| DLBC | |||
| PCPG | |||
| THCA | |||
| THYM | |||
| UCS |
| Cancer | P-value | Q-value |
|---|---|---|
| THYM | ||
| KIRC | ||
| ACC | ||
| LUSC | ||
| PCPG | ||
| CESC | ||
| READ | ||
| LAML | ||
| LIHC | ||
| DLBC | ||
| LGG | ||
| THCA | ||
| LUAD |